2MPT
 
 | | WW3 domain of Nedd4L in complex with its HECT domain PY motif | | 分子名称: | E3 ubiquitin-protein ligase NEDD4-like | | 著者 | Escobedo, A, Macias, M.J, Gomes, T, Aragon, E, Martin-Malpartida, P, Ruiz, L. | | 登録日 | 2014-06-02 | | 公開日 | 2014-10-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structural Basis of the Activation and Degradation Mechanisms of the E3 Ubiquitin Ligase Nedd4L.
Structure, 22, 2014
|
|
2LTV
 
 | | YAP WW2 in complex with a Smad7 derived peptide | | 分子名称: | Smad7 derived peptide, Yorkie homolog | | 著者 | Macias, M.J, Aragon, E, Goerner, N, Xi, Q, Lopes, T, Gao, S, Massague, J. | | 登録日 | 2012-06-04 | | 公開日 | 2012-11-21 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structural Basis for the Versatile Interactions of Smad7 with Regulator WW Domains in TGF-beta Pathways.
Structure, 20, 2012
|
|
6FZS
 
 | |
6FZT
 
 | | Crystal structure of Smad8_9-MH1 bound to the GGCGC site. | | 分子名称: | DNA (5'-D(P*TP*GP*CP*AP*GP*GP*CP*GP*CP*GP*CP*CP*TP*GP*CP*A)-3'), GLYCEROL, Mothers against decapentaplegic homolog 9, ... | | 著者 | Kaczmarska, Z, Marquez, J.A, Macias, M.J. | | 登録日 | 2018-03-15 | | 公開日 | 2019-10-02 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.46 Å) | | 主引用文献 | Unveiling the dimer/monomer propensities of Smad MH1-DNA complexes
Computational and Structural Biotechnology Journal, 19, 2021
|
|
6H3R
 
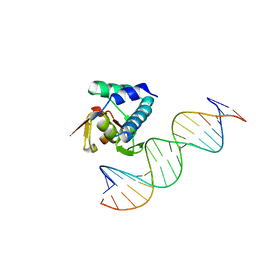 | |
5OD6
 
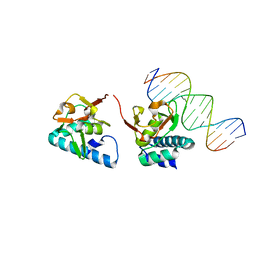 | |
5ODG
 
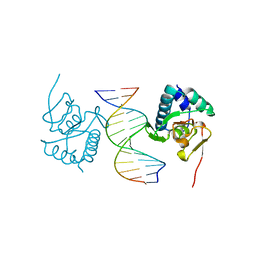 | | Crystal structure of Smad3-MH1 bound to the GGCT site. | | 分子名称: | CHLORIDE ION, DNA (5'-D(P*CP*AP*GP*GP*CP*TP*AP*GP*CP*CP*TP*GP*CP*A)-3'), Mothers against decapentaplegic homolog 3, ... | | 著者 | Kaczmarska, Z, Marquez, J.A, Macias, M.J. | | 登録日 | 2017-07-05 | | 公開日 | 2017-11-15 | | 最終更新日 | 2017-12-27 | | 実験手法 | X-RAY DIFFRACTION (2.12 Å) | | 主引用文献 | Structural basis for genome wide recognition of 5-bp GC motifs by SMAD transcription factors.
Nat Commun, 8, 2017
|
|
5MEZ
 
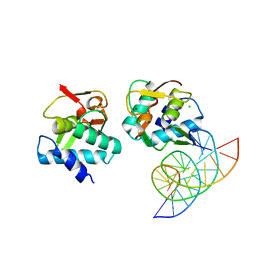 | | Crystal structure of Smad4-MH1 bound to the GGCT site. | | 分子名称: | CHLORIDE ION, DNA (5'-D(P*GP*CP*AP*GP*GP*CP*TP*AP*GP*CP*CP*TP*GP*CP*A)-3'), MH1 domain of human Smad4, ... | | 著者 | Kaczmarska, Z, Freier, R, Marquez, J.A, Macias, M.J. | | 登録日 | 2016-11-16 | | 公開日 | 2017-11-15 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.98 Å) | | 主引用文献 | Structural basis for genome wide recognition of 5-bp GC motifs by SMAD transcription factors.
Nat Commun, 8, 2017
|
|
6ZMN
 
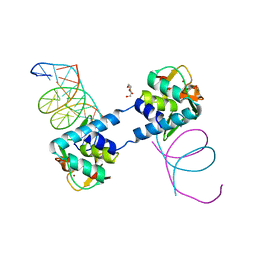 | |
5MEY
 
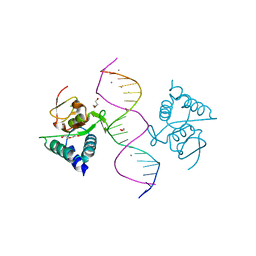 | | Crystal structure of Smad4-MH1 bound to the GGCGC site. | | 分子名称: | 1,2-ETHANEDIOL, CALCIUM ION, CHLORIDE ION, ... | | 著者 | Kaczmarska, Z, Freier, R, Marquez, J.A, Macias, M.J. | | 登録日 | 2016-11-16 | | 公開日 | 2017-11-15 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.05 Å) | | 主引用文献 | Structural basis for genome wide recognition of 5-bp GC motifs by SMAD transcription factors.
Nat Commun, 8, 2017
|
|
5MF0
 
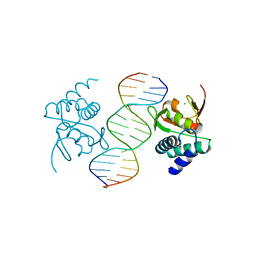 | | Crystal structure of Smad4-MH1 bound to the GGCCG site. | | 分子名称: | CHLORIDE ION, DNA (5'-D(P*AP*CP*GP*GP*GP*CP*CP*GP*CP*GP*GP*CP*CP*CP*GP*T)-3'), MH1 domain of human Smad4, ... | | 著者 | Kaczmarska, Z, Freier, R, Marquez, J.A, Macias, M.J. | | 登録日 | 2016-11-16 | | 公開日 | 2017-11-15 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (3.03 Å) | | 主引用文献 | Structural basis for genome wide recognition of 5-bp GC motifs by SMAD transcription factors.
Nat Commun, 8, 2017
|
|
5NM9
 
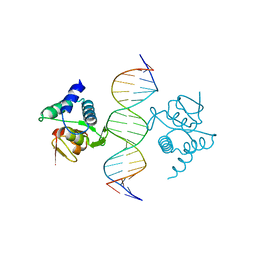 | | Crystal structure of the placozoa Trichoplax adhaerens Smad4-MH1 bound to the GGCGC site. | | 分子名称: | DNA (5'-D(P*AP*TP*GP*CP*GP*GP*GP*CP*GP*CP*GP*CP*CP*CP*GP*CP*AP*T)-3'), Mothers against decapentaplegic homolog, ZINC ION | | 著者 | Kaczmarska, Z, Freier, R, Marquez, J.A, Macias, M.J. | | 登録日 | 2017-04-05 | | 公開日 | 2017-11-15 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.43 Å) | | 主引用文献 | Structural basis for genome wide recognition of 5-bp GC motifs by SMAD transcription factors.
Nat Commun, 8, 2017
|
|
6TBZ
 
 | |
6TCE
 
 | |
2LTW
 
 | | YAP WW1 in complex with a Smad7 derived peptide | | 分子名称: | Smad7 derived peptide, Yorkie homolog | | 著者 | Macias, M.J, Aragon, E, Goerner, N, Xi, Q, Lopes, T, Gao, S, Massague, J. | | 登録日 | 2012-06-04 | | 公開日 | 2012-11-21 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structural Basis for the Versatile Interactions of Smad7 with Regulator WW Domains in TGF-beta Pathways.
Structure, 20, 2012
|
|
2LTX
 
 | | Smurf1 WW2 domain in complex with a Smad7 derived peptide | | 分子名称: | E3 ubiquitin-protein ligase SMURF1, Smad7 derived peptide | | 著者 | Macias, M.J, Aragon, E, Goerner, N, Xi, Q, Lopes, T, Gao, S, Massague, J. | | 登録日 | 2012-06-04 | | 公開日 | 2012-11-21 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structural Basis for the Versatile Interactions of Smad7 with Regulator WW Domains in TGF-beta Pathways.
Structure, 20, 2012
|
|
2LTY
 
 | | NEDD4L WW2 domain in complex with a Smad7 derived peptide | | 分子名称: | E3 ubiquitin-protein ligase NEDD4-like, Smad7 derived peptide | | 著者 | Macias, M.J, Aragon, E, Goerner, N, Xi, Q, Lopes, T, Gao, S, Massague, J. | | 登録日 | 2012-06-04 | | 公開日 | 2012-11-21 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structural Basis for the Versatile Interactions of Smad7 with Regulator WW Domains in TGF-beta Pathways.
Structure, 20, 2012
|
|
2LTZ
 
 | | Smurf2 WW3 domain in complex with a Smad7 derived peptide | | 分子名称: | E3 ubiquitin-protein ligase SMURF2, Smad7 derived peptide | | 著者 | Macias, M.J, Aragon, E, Goerner, N, Xi, Q, Lopes, T, Gao, S, Massague, J. | | 登録日 | 2012-06-04 | | 公開日 | 2012-11-21 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structural Basis for the Versatile Interactions of Smad7 with Regulator WW Domains in TGF-beta Pathways.
Structure, 20, 2012
|
|
