3BIK
 
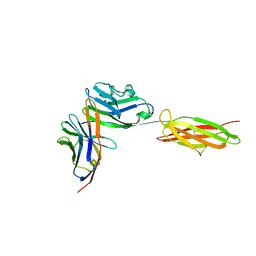 | | Crystal Structure of the PD-1/PD-L1 Complex | | Descriptor: | GLYCEROL, Programmed cell death 1 ligand 1, Programmed cell death protein 1 | | Authors: | Lin, D.Y, Tanaka, Y, Iwasaki, M, Gittis, A.G, Su, H.P, Mikami, B, Okazaki, T, Honjo, T, Minato, N, Garboczi, D.N. | | Deposit date: | 2007-11-30 | | Release date: | 2008-02-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | The PD-1/PD-L1 complex resembles the antigen-binding Fv domains of antibodies and T cell receptors.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
3BIS
 
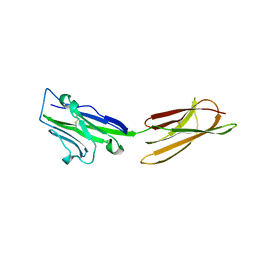 | | Crystal Structure of the PD-L1 | | Descriptor: | Programmed cell death 1 ligand 1 | | Authors: | Lin, D.Y, Tanaka, Y, Iwasaki, M, Gittis, A.G, Su, H.P, Mikami, B, Okazaki, T, Honjo, T, Minato, N, Garboczi, D.N. | | Deposit date: | 2007-11-30 | | Release date: | 2008-02-26 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | The PD-1/PD-L1 complex resembles the antigen-binding Fv domains of antibodies and T cell receptors.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
3C64
 
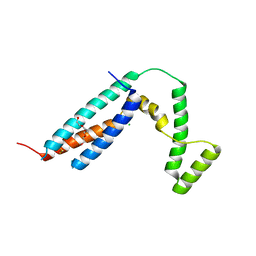 | | The MC179 portion of the Cysteine-rich Interdomain Region (CIDR) of a Plasmodium falciparum Erythrocyte Membrane Protein-1 (PfEMP1) | | Descriptor: | CHLORIDE ION, PfEMP1 variant 2 of strain MC, TETRAETHYLENE GLYCOL | | Authors: | Klein, M.M, Gittis, A.G, Su, H.P, Makobongo, M.O, Moore, J.M, Singh, S, Miller, L.H, Garboczi, D.N. | | Deposit date: | 2008-02-02 | | Release date: | 2008-09-16 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The Cysteine-Rich Interdomain Region from the Highly Variable Plasmodium falciparum Erythrocyte Membrane Protein-1 Exhibits a Conserved Structure.
Plos Pathog., 4, 2008
|
|
3CPZ
 
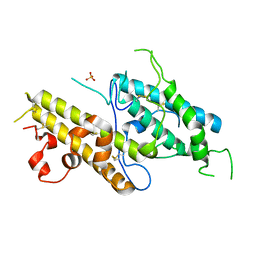 | | Crystal structure of VAR2CSA DBL3x domain in the presence of dodecasaccharide of CSA | | Descriptor: | Erythrocyte membrane protein 1, SULFATE ION | | Authors: | Singh, K, Gittis, A.G, Nguyen, P, Gowda, D.C, Miller, L.H, Garboczi, D.N. | | Deposit date: | 2008-04-01 | | Release date: | 2008-09-02 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of the DBL3x domain of pregnancy-associated malaria protein VAR2CSA complexed with chondroitin sulfate A.
Nat.Struct.Mol.Biol., 15, 2008
|
|
3CML
 
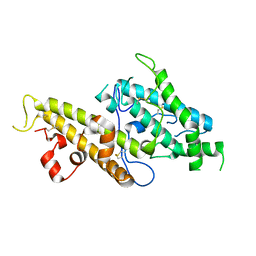 | | Crystal Structure of the DBL3x domain of the Plasmodium falcipurum VAR2CSA protein | | Descriptor: | Erythrocyte membrane protein 1 | | Authors: | Singh, K, Gittis, A.G, Nguyen, P, Gowda, D.C, Miller, L.H, Garboczi, D.N. | | Deposit date: | 2008-03-23 | | Release date: | 2008-09-02 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of the DBL3x domain of pregnancy-associated malaria protein VAR2CSA complexed with chondroitin sulfate A.
Nat.Struct.Mol.Biol., 15, 2008
|
|
1MUT
 
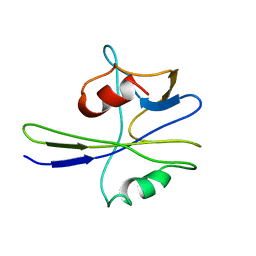 | | NMR STUDY OF MUTT ENZYME, A NUCLEOSIDE TRIPHOSPHATE PYROPHOSPHOHYDROLASE | | Descriptor: | NUCLEOSIDE TRIPHOSPHATE PYROPHOSPHOHYDROLASE | | Authors: | Abeygunawardana, C, Weber, D.J, Gittis, A.G, Frick, D.N, Lin, J, Miller, A.-F, Bessman, M.J, Mildvan, A.S. | | Deposit date: | 1995-09-14 | | Release date: | 1996-04-03 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the MutT enzyme, a nucleoside triphosphate pyrophosphohydrolase.
Biochemistry, 34, 1995
|
|
1QA6
 
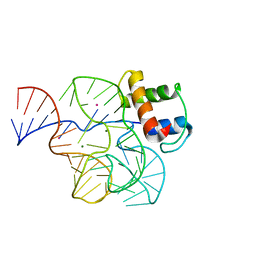 | | CRYSTAL STRUCTURE OF A CONSERVED RIBOSOMAL PROTEIN-RNA COMPLEX | | Descriptor: | 58 NUCLEOTIDE RIBOSOMAL RNA DOMAIN, MAGNESIUM ION, OSMIUM ION, ... | | Authors: | Conn, G.L, Draper, D.E, Lattman, E.E, Gittis, A.G. | | Deposit date: | 1999-04-15 | | Release date: | 1999-05-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of a conserved ribosomal protein-RNA complex.
Science, 284, 1999
|
|
2OF1
 
 | |
2OXP
 
 | |
2SOB
 
 | | SN-OB, OB-FOLD SUB-DOMAIN OF STAPHYLOCOCCAL NUCLEASE, NMR, 10 STRUCTURES | | Descriptor: | STAPHYLOCOCCAL NUCLEASE | | Authors: | Alexandrescu, A.T, Gittis, A.G, Abeygunawardana, C, Shortle, D. | | Deposit date: | 1995-09-15 | | Release date: | 1995-12-07 | | Last modified: | 2021-11-03 | | Method: | SOLUTION NMR | | Cite: | NMR structure of a stable "OB-fold" sub-domain isolated from staphylococcal nuclease.
J.Mol.Biol., 250, 1995
|
|
2SNM
 
 | | IN A STAPHYLOCOCCAL NUCLEASE MUTANT THE SIDE-CHAIN of A LYSINE REPLACING VALINE 66 IS FULLY BURIED IN THE HYDROPHOBIC CORE | | Descriptor: | CALCIUM ION, STAPHYLOCOCCAL NUCLEASE, THYMIDINE-3',5'-DIPHOSPHATE | | Authors: | Stites, W.E, Gittis, A.G, Lattman, E.E, Shortle, D. | | Deposit date: | 1991-04-03 | | Release date: | 1993-01-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | In a staphylococcal nuclease mutant the side-chain of a lysine replacing valine 66 is fully buried in the hydrophobic core.
J.Mol.Biol., 221, 1991
|
|
2ENB
 
 | |
1ENC
 
 | | CRYSTAL STRUCTURES OF THE BINARY CA2+ AND PDTP COMPLEXES AND THE TERNARY COMPLEX OF THE ASP 21->GLU MUTANT OF STAPHYLOCOCCAL NUCLEASE. IMPLICATIONS FOR CATALYSIS AND LIGAND BINDING | | Descriptor: | CALCIUM ION, STAPHYLOCOCCAL NUCLEASE, THYMIDINE-3',5'-DIPHOSPHATE | | Authors: | Libson, A, Gittis, A, Lattman, E.E. | | Deposit date: | 1994-02-14 | | Release date: | 1994-05-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structures of the binary Ca2+ and pdTp complexes and the ternary complex of the Asp21-->Glu mutant of staphylococcal nuclease. Implications for catalysis and ligand binding.
Biochemistry, 33, 1994
|
|
1ENA
 
 | |
3K7B
 
 | |
7TVY
 
 | |
7TVC
 
 | | Short form D7 protein from Aedes aegypti | | Descriptor: | Salivary short D7 protein | | Authors: | Andersen, J.F, Xu, X. | | Deposit date: | 2022-02-04 | | Release date: | 2022-05-25 | | Method: | X-RAY DIFFRACTION (1.19 Å) | | Cite: | Functional aspects of evolution in a cluster of salivary protein genes from mosquitoes.
Insect Biochem.Mol.Biol., 146, 2022
|
|
7TX8
 
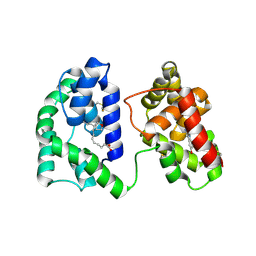 | | Long form D7 protein from Anopheles darlingi with U46619 and serotonin bound | | Descriptor: | (5Z)-7-{(1R,4S,5S,6R)-6-[(1E,3S)-3-hydroxyoct-1-en-1-yl]-2-oxabicyclo[2.2.1]hept-5-yl}hept-5-enoic acid, Long form D7 salivary protein, SEROTONIN | | Authors: | Andersen, J.F, Alvarenga, P.H. | | Deposit date: | 2022-02-08 | | Release date: | 2022-06-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Functional aspects of evolution in a cluster of salivary protein genes from mosquitoes.
Insect Biochem.Mol.Biol., 146, 2022
|
|
1TUM
 
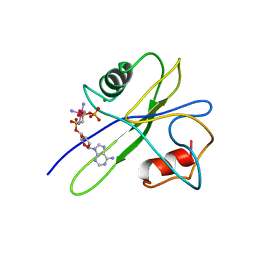 | | MUTT PYROPHOSPHOHYDROLASE-METAL-NUCLEOTIDE-METAL COMPLEX, NMR, 16 STRUCTURES | | Descriptor: | COBALT TETRAAMMINE ION, DIPHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER, MAGNESIUM ION, ... | | Authors: | Lin, J, Abeygunawardana, C, Frick, D.N, Bessman, M.J, Mildvan, A.S. | | Deposit date: | 1996-12-05 | | Release date: | 1997-05-15 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the quaternary MutT-M2+-AMPCPP-M2+ complex and mechanism of its pyrophosphohydrolase action.
Biochemistry, 36, 1997
|
|
1PPX
 
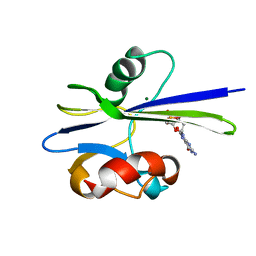 | | Solution Structure of the MutT Pyrophosphohydrolase Complexed with Mg(2+) and 8-oxo-dGMP, a Tightly-bound Product | | Descriptor: | 8-OXO-2'-DEOXY-GUANOSINE-5'-MONOPHOSPHATE, MAGNESIUM ION, Mutator mutT protein | | Authors: | Massiah, M.A, Saraswat, V, Azurmendi, H.F, Mildvan, A.S. | | Deposit date: | 2003-06-17 | | Release date: | 2003-08-26 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Solution structure and NH exchange studies of the MutT pyrophosphohydrolase complexed with Mg(2+) and 8-oxo-dGMP, a tightly bound product.
Biochemistry, 42, 2003
|
|
1PUN
 
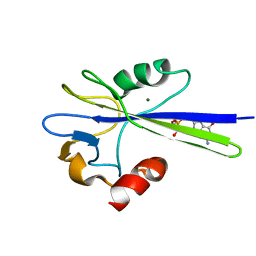 | | Solution Structure of the MutT Pyrophosphohydrolase Complexed with Mg(2+) and 8-oxo-dGMP, a Tightly-bound Product | | Descriptor: | 8-OXO-2'-DEOXY-GUANOSINE-5'-MONOPHOSPHATE, MAGNESIUM ION, Mutator mutT protein | | Authors: | Massiah, M.A, Saraswat, V, Azurmendi, H.F, Mildvan, A.S. | | Deposit date: | 2003-06-25 | | Release date: | 2003-08-26 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Solution structure and NH exchange studies of the MutT pyrophosphohydrolase complexed with Mg(2+) and 8-oxo-dGMP, a tightly bound product.
Biochemistry, 42, 2003
|
|
1PUQ
 
 | | Solution Structure of the MutT Pyrophosphohydrolase Complexed with Mg(2+) and 8-oxo-dGMP, a Tightly-bound Product | | Descriptor: | 8-OXO-2'-DEOXY-GUANOSINE-5'-MONOPHOSPHATE, MAGNESIUM ION, Mutator mutT protein | | Authors: | Massiah, M.A, Saraswat, V, Azurmendi, H.F, Mildvan, A.S. | | Deposit date: | 2003-06-25 | | Release date: | 2003-08-26 | | Last modified: | 2019-11-20 | | Method: | SOLUTION NMR | | Cite: | Solution structure and NH exchange studies of the MutT pyrophosphohydrolase complexed with Mg(2+) and 8-oxo-dGMP, a tightly bound product.
Biochemistry, 42, 2003
|
|
1PUS
 
 | | Solution Structure of the MutT Pyrophosphohydrolase Complexed with Mg(2+) and 8-oxo-dGMP, a Tightly-bound Product | | Descriptor: | 8-OXO-2'-DEOXY-GUANOSINE-5'-MONOPHOSPHATE, MAGNESIUM ION, Mutator mutT protein | | Authors: | Massiah, M.A, Saraswat, V, Azurmendi, H.F, Mildvan, A.S. | | Deposit date: | 2003-06-25 | | Release date: | 2003-08-26 | | Last modified: | 2019-11-20 | | Method: | SOLUTION NMR | | Cite: | Solution structure and NH exchange studies of the MutT pyrophosphohydrolase complexed with Mg(2+) and 8-oxo-dGMP, a tightly bound product.
Biochemistry, 42, 2003
|
|
1OLN
 
 | | Model for thiostrepton antibiotic binding to L11 substrate from 50S ribosomal RNA | | Descriptor: | 50S RIBOSOMAL PROTEIN L11, RNA, THIOSTREPTON | | Authors: | Lentzen, G, Klinck, R, Matassova, N, Aboul-Ela, F, Murchie, A.I.H. | | Deposit date: | 2003-08-08 | | Release date: | 2003-09-11 | | Last modified: | 2019-08-21 | | Method: | SOLUTION NMR, THEORETICAL MODEL | | Cite: | Structural Basis for Contrasting Activities of Ribosome Binding Thiazole Antibiotics
Chem.Biol., 10, 2003
|
|
