8OST
 
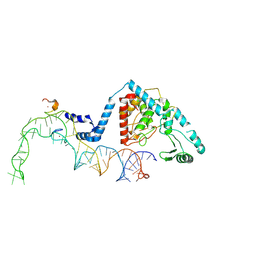 | | Structure of human terminal uridylyltransferase 4 (TUT4, ZCCHC11) in complex with pre-let7g miRNA and Lin28A | | Descriptor: | Protein lin-28 homolog A, Terminal uridylyltransferase 4, ZINC ION, ... | | Authors: | Gilbert, R.J, Yi, G, Ye, M. | | Deposit date: | 2023-04-20 | | Release date: | 2024-07-17 | | Last modified: | 2024-09-25 | | Method: | ELECTRON MICROSCOPY (3.69 Å) | | Cite: | Structural basis for activity switching in polymerases determining the fate of let-7 pre-miRNAs.
Nat.Struct.Mol.Biol., 31, 2024
|
|
4E80
 
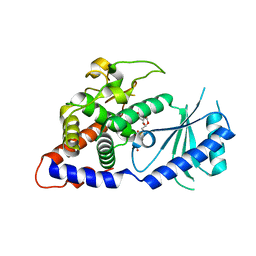 | | Structural Basis for the Activity of a Cytoplasmic RNA Terminal U-transferase | | Descriptor: | Poly(A) RNA polymerase protein cid1, URIDINE 5'-TRIPHOSPHATE | | Authors: | Yates, L.A, Fleurdepine, S, Rissland, O.S, DeColibus, L, Harlos, K, Norbury, C.J, Gilbert, R.J.C. | | Deposit date: | 2012-03-19 | | Release date: | 2012-07-04 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.02 Å) | | Cite: | Structural basis for the activity of a cytoplasmic RNA terminal uridylyl transferase.
Nat.Struct.Mol.Biol., 19, 2012
|
|
4E8F
 
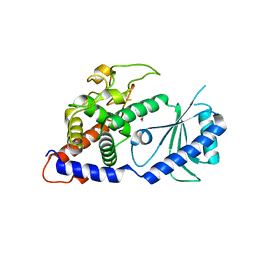 | | Structural Basis for the Activity of a Cytoplasmic RNA Terminal U-transferase | | Descriptor: | ACETATE ION, GLYCEROL, Poly(A) RNA polymerase protein cid1 | | Authors: | Yates, L.A, Fleurdepine, S, Rissland, O.S, DeColibus, L, Harlos, K, Norbury, C.J, Gilbert, R.J.C. | | Deposit date: | 2012-03-20 | | Release date: | 2012-07-04 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis for the activity of a cytoplasmic RNA terminal uridylyl transferase.
Nat.Struct.Mol.Biol., 19, 2012
|
|
4E7X
 
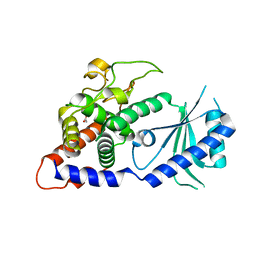 | | Structural Basis for the Activity of a Cytoplasmic RNA Terminal U-transferase | | Descriptor: | ACETATE ION, Poly(A) RNA polymerase protein cid1 | | Authors: | Yates, L.A, Fleurdepine, S, Rissland, O.S, DeColibus, L, Harlos, K, Norbury, C.J, Gilbert, R.J.C. | | Deposit date: | 2012-03-19 | | Release date: | 2012-07-04 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural basis for the activity of a cytoplasmic RNA terminal uridylyl transferase.
Nat.Struct.Mol.Biol., 19, 2012
|
|
4UD4
 
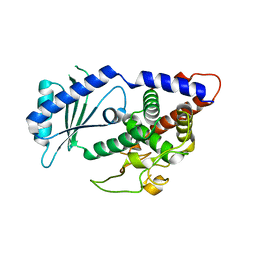 | | Structural Plasticity of Cid1 Provides a Basis for its RNA Terminal Uridylyl Transferase Activity | | Descriptor: | GLYCEROL, POLY(A) RNA POLYMERASE PROTEIN CID1 | | Authors: | Yates, L.A, Durrant, B.P, Fleurdepine, S, Harlos, K, Norbury, C.J, Gilbert, R.J.C. | | Deposit date: | 2014-12-07 | | Release date: | 2015-03-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Structural plasticity of Cid1 provides a basis for its distributive RNA terminal uridylyl transferase activity.
Nucleic Acids Res., 43, 2015
|
|
5L5N
 
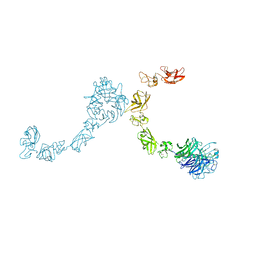 | | Plexin A4 full extracellular region, domains 1 to 7 modeled, data to 8.5 angstrom, spacegroup P4(3)22 | | Descriptor: | Plexin-A4 | | Authors: | Janssen, B.J.C, Kong, Y, Malinauskas, T, Vangoor, V.R, Coles, C.H, Kaufmann, R, Ni, T, Gilbert, R.J.C, Padilla-Parra, S, Pasterkamp, R.J, Jones, E.Y. | | Deposit date: | 2016-05-28 | | Release date: | 2016-07-06 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (8.502 Å) | | Cite: | Structural Basis for Plexin Activation and Regulation.
Neuron, 91, 2016
|
|
5L5G
 
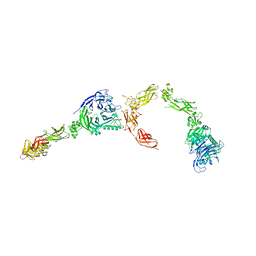 | | Plexin A2 full extracellular region, domains 1 to 8 modeled, data to 10 angstrom | | Descriptor: | Plexin-A2 | | Authors: | Janssen, B.J.C, Kong, Y, Malinauskas, T, Vangoor, V.R, Coles, C.H, Kaufmann, R, Ni, T, Gilbert, R.J.C, Padilla-Parra, S, Pasterkamp, R.J, Jones, E.Y. | | Deposit date: | 2016-05-28 | | Release date: | 2016-07-06 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (10 Å) | | Cite: | Structural Basis for Plexin Activation and Regulation.
Neuron, 91, 2016
|
|
5L5M
 
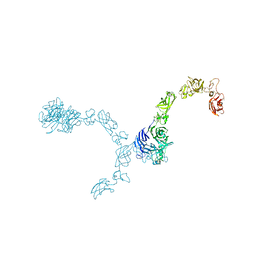 | | Plexin A4 full extracellular region, domains 1 to 7 modeled, data to 8 angstrom, spacegroup P4(3)2(1)2 | | Descriptor: | Plexin-A4 | | Authors: | Janssen, B.J.C, Kong, Y, Malinauskas, T, Vangoor, V.R, Coles, C.H, Kaufmann, R, Ni, T, Gilbert, R.J.C, Padilla-Parra, S, Pasterkamp, R.J, Jones, E.Y. | | Deposit date: | 2016-05-28 | | Release date: | 2016-07-06 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (8 Å) | | Cite: | Structural Basis for Plexin Activation and Regulation.
Neuron, 91, 2016
|
|
5L59
 
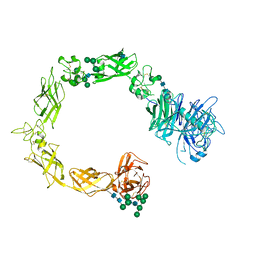 | | Plexin A1 full extracellular region, domains 1 to 10, to 6 angstrom, spacegroup P2(1) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Plexin-A1, alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Janssen, B.J.C, Kong, Y, Malinauskas, T, Vangoor, V.R, Coles, C.H, Kaufmann, R, Ni, T, Gilbert, R.J.C, Padilla-Parra, S, Pasterkamp, R.J, Jones, E.Y. | | Deposit date: | 2016-05-28 | | Release date: | 2016-07-06 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (6 Å) | | Cite: | Structural Basis for Plexin Activation and Regulation.
Neuron, 91, 2016
|
|
5L5K
 
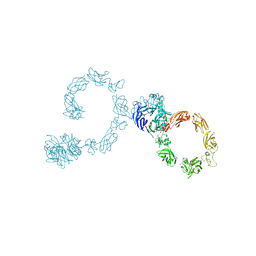 | | Plexin A4 full extracellular region, domains 1 to 10, data to 7.5 angstrom, spacegroup P4(1) | | Descriptor: | Plexin-A4 | | Authors: | Janssen, B.J.C, Kong, Y, Malinauskas, T, Vangoor, V.R, Coles, C.H, Kaufmann, R, Ni, T, Gilbert, R.J.C, Padilla-Parra, S, Pasterkamp, R.J, Jones, E.Y. | | Deposit date: | 2016-05-28 | | Release date: | 2016-07-06 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (7.501 Å) | | Cite: | Structural Basis for Plexin Activation and Regulation.
Neuron, 91, 2016
|
|
5L5L
 
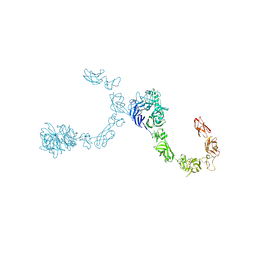 | | Plexin A4 full extracellular region, domains 1 to 8 modeled, data to 8 angstrom, spacegroup P2(1) | | Descriptor: | Plexin-A4 | | Authors: | Janssen, B.J.C, Kong, Y, Malinauskas, T, Vangoor, V.R, Coles, C.H, Kaufmann, R, Ni, T, Gilbert, R.J.C, Padilla-Parra, S, Pasterkamp, R.J, Jones, E.Y. | | Deposit date: | 2016-05-28 | | Release date: | 2016-07-06 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (8.001 Å) | | Cite: | Structural Basis for Plexin Activation and Regulation.
Neuron, 91, 2016
|
|
5L56
 
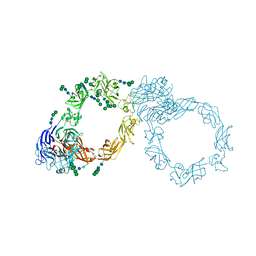 | | Plexin A1 full extracellular region, domains 1 to 10, to 4 angstrom | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Plexin-A1, alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Janssen, B.J.C, Kong, Y, Malinauskas, T, Vangoor, V.R, Coles, C.H, Kaufmann, R, Ni, T, Gilbert, R.J.C, Padilla-Parra, S, Pasterkamp, R.J, Jones, E.Y. | | Deposit date: | 2016-05-28 | | Release date: | 2016-07-06 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Structural Basis for Plexin Activation and Regulation.
Neuron, 91, 2016
|
|
5L74
 
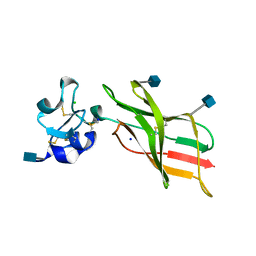 | | Plexin A2 extracellular segment domains 4-5 (PSI2-IPT2), resolution 1.36 Angstrom | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, Plexin-A2, ... | | Authors: | Kong, Y, Janssen, B.J.C, Malinauskas, T, Vangoor, V.R, Coles, C.H, Kaufmann, R, Ni, T, Gilbert, R.J.C, Padilla-Parra, S, Pasterkamp, R.J, Jones, E.Y. | | Deposit date: | 2016-06-01 | | Release date: | 2017-03-15 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Structural Basis for Plexin Activation and Regulation.
Neuron, 91, 2016
|
|
5L7N
 
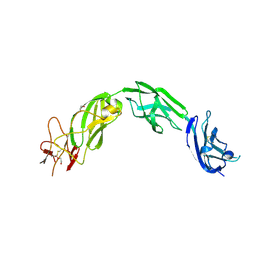 | | Plexin A1 extracellular fragment, domains 7-10 (IPT3-IPT6) | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Kong, Y, Janssen, B.J.C, Malinauskas, T, Vangoor, V.R, Coles, C.H, Kaufmann, R, Ni, T, Gilbert, R.J.C, Padilla-Parra, S, Pasterkamp, R.J, Jones, E.Y. | | Deposit date: | 2016-06-03 | | Release date: | 2017-03-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Basis for Plexin Activation and Regulation.
Neuron, 91, 2016
|
|
5L5C
 
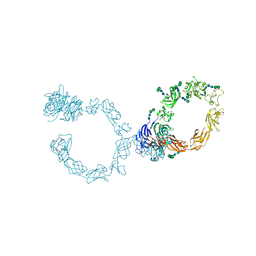 | | Plexin A1 full extracellular region, domains 1 to 10, to 6 angstrom, spacegroup P4(3)2(1)2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Plexin-A1, ... | | Authors: | Janssen, B.J.C, Kong, Y, Malinauskas, T, Vangoor, V.R, Coles, C.H, Kaufmann, R, Ni, T, Gilbert, R.J.C, Padilla-Parra, S, Pasterkamp, R.J, Jones, E.Y. | | Deposit date: | 2016-05-28 | | Release date: | 2016-07-06 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (6 Å) | | Cite: | Structural Basis for Plexin Activation and Regulation.
Neuron, 91, 2016
|
|
7PLN
 
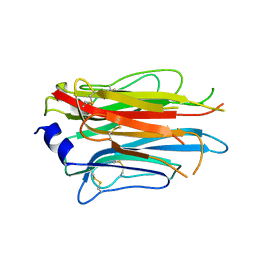 | | Structure of the APCbeta domain of Plasmodium vivax perforin-like protein 1 | | Descriptor: | Sporozoite micronemal protein essential for cell traversal, putative | | Authors: | Williams, S.I, Ni, T, Yu, X, Gilbert, R.J.C. | | Deposit date: | 2021-08-31 | | Release date: | 2022-06-29 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Structural, Functional and Computational Studies of Membrane Recognition by Plasmodium Perforin-Like Proteins 1 and 2
J.Mol.Biol., 434, 2022
|
|
5FMV
 
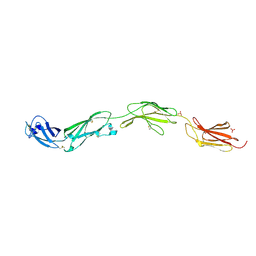 | | Crystal structure of human CD45 extracellular region, domains d1-d4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, RECEPTOR-TYPE TYROSINE-PROTEIN PHOSPHATASE C, SULFATE ION | | Authors: | Chang, V.T, Fernandes, R.A, Ganzinger, K.A, Lee, S.F, Siebold, C, McColl, J, Jonsson, P, Palayret, M, Harlos, K, Coles, C.H, Jones, E.Y, Lui, Y, Huang, E, Gilbert, R.J.C, Klenerman, D, Aricescu, A.R, Davis, S.J. | | Deposit date: | 2015-11-09 | | Release date: | 2016-03-23 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Initiation of T Cell Signaling by Cd45 Segregation at 'Close Contacts'.
Nat.Immunol., 17, 2016
|
|
5FN6
 
 | | Crystal structure of human CD45 extracellular region, domains d1-d3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, RECEPTOR-TYPE TYROSINE-PROTEIN PHOSPHATASE C | | Authors: | Chang, V.T, Fernandes, R.A, Ganzinger, K.A, Lee, S.F, Siebold, C, McColl, J, Jonsson, P, Palayret, M, Harlos, K, Coles, C.H, Jones, E.Y, Lui, Y, Huang, E, Gilbert, R.J.C, Klenerman, D, Aricescu, A.R, Davis, S.J. | | Deposit date: | 2015-11-10 | | Release date: | 2016-03-23 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Initiation of T Cell Signaling by Cd45 Segregation at 'Close Contacts'.
Nat.Immunol., 17, 2016
|
|
5FN8
 
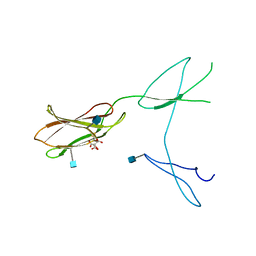 | | Crystal structure of rat CD45 extracellular region, domains d3-d4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CITRATE ANION, RECEPTOR-TYPE TYROSINE-PROTEIN PHOSPHATASE C | | Authors: | Chang, V.T, Fernandes, R.A, Ganzinger, K.A, Lee, S.F, Siebold, C, McColl, J, Jonsson, P, Palayret, M, Harlos, K, Coles, C.H, Jones, E.Y, Lui, Y, Huang, E, Gilbert, R.J.C, Klenerman, D, Aricescu, A.R, Davis, S.J. | | Deposit date: | 2015-11-11 | | Release date: | 2016-03-23 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Initiation of T Cell Signaling by Cd45 Segregation at 'Close Contacts'.
Nat.Immunol., 17, 2016
|
|
5FN7
 
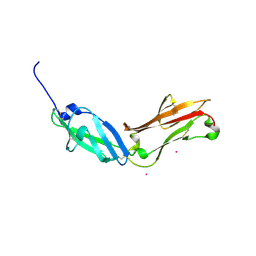 | | Crystal structure of human CD45 extracellular region, domains d1-d2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, MERCURY (II) ION, RECEPTOR-TYPE TYROSINE-PROTEIN PHOSPHATASE C | | Authors: | Chang, V.T, Fernandes, R.A, Ganzinger, K.A, Lee, S.F, Siebold, C, McColl, J, Jonsson, P, Palayret, M, Harlos, K, Coles, C.H, Jones, E.Y, Lui, Y, Huang, E, Gilbert, R.J.C, Klenerman, D, Aricescu, A.R, Davis, S.J. | | Deposit date: | 2015-11-10 | | Release date: | 2016-03-23 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Initiation of T Cell Signaling by Cd45 Segregation at 'Close Contacts'.
Nat.Immunol., 17, 2016
|
|
8OEF
 
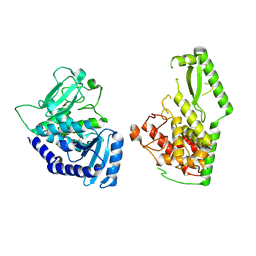 | |
8OPS
 
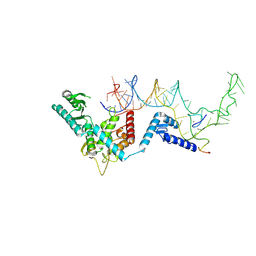 | | Human terminal uridylyltransferase 7 (TUT7/ZCCHC6) bound with pre-let7g miRNA and Lin28A - complex 1 | | Descriptor: | Protein lin-28 homolog A, RNA (71-MER) Let7g, Terminal uridylyltransferase 7, ... | | Authors: | Yi, G, Ye, M, Gilbert, R.J. | | Deposit date: | 2023-04-08 | | Release date: | 2024-07-24 | | Last modified: | 2024-09-25 | | Method: | ELECTRON MICROSCOPY (3.82 Å) | | Cite: | Structural basis for activity switching in polymerases determining the fate of let-7 pre-miRNAs.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8OPP
 
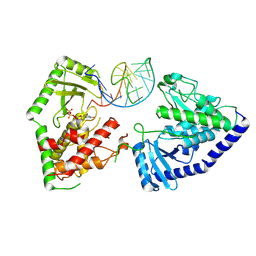 | | Structure of human terminal uridylyltransferase 7 (hTUT7/ZCCHC6) bound with pre-let7g miRNA and UTPalphaS | | Descriptor: | RNA (25-MER), Terminal uridylyltransferase 7, [[(2~{R},3~{S},4~{R},5~{R})-5-[2,4-bis(oxidanylidene)pyrimidin-1-yl]-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-sulfanyl-phosphoryl] phosphono hydrogen phosphate | | Authors: | Yi, G, Ye, M, Gilbert, R.J. | | Deposit date: | 2023-04-07 | | Release date: | 2024-07-24 | | Last modified: | 2024-09-25 | | Method: | ELECTRON MICROSCOPY (3.76 Å) | | Cite: | Structural basis for activity switching in polymerases determining the fate of let-7 pre-miRNAs.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8OPT
 
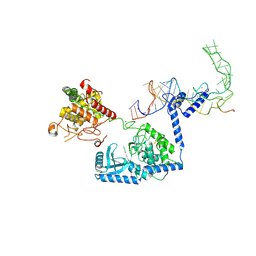 | | Human terminal uridylyltransferase 7 (TUT7/ZCCHC6) bound with pre-let7g miRNA and Lin28A - complex 2 | | Descriptor: | Protein lin-28 homolog A, RNA (53-MER), Terminal uridylyltransferase 7, ... | | Authors: | Yi, G, Ye, M, Gilbert, R.J. | | Deposit date: | 2023-04-08 | | Release date: | 2024-07-24 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.65 Å) | | Cite: | Structural basis for activity switching in polymerases determining the fate of let-7 pre-miRNAs.
Nat.Struct.Mol.Biol., 31, 2024
|
|
4BQ7
 
 | | Crystal structure of the RGMB-Neo1 complex form 2 | | Descriptor: | NEOGENIN, RGM DOMAIN FAMILY MEMBER B | | Authors: | Bell, C.H, Healey, E, van Erp, S, Bishop, B, Tang, C, Gilbert, R.J.C, Aricescu, A.R, Pasterkamp, R.J, Siebold, C. | | Deposit date: | 2013-05-30 | | Release date: | 2013-06-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (6.601 Å) | | Cite: | Structure of the Repulsive Guidance Molecule (Rgm)-Neogenin Signaling Hub
Science, 341, 2013
|
|
