7LHO
 
 | |
7LHM
 
 | |
8DN1
 
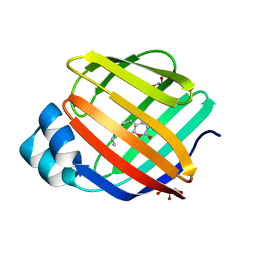 | | Q108K:K40L:T51C:T53A:R58L:Q38F:Q4F mutant of hCRBPII bound to synthetic fluorophore CM1V at pH 7.2 | | Descriptor: | (2E)-3-[7-(diethylamino)-2-oxo-2H-1-benzopyran-3-yl]prop-2-enal, bound form, GLYCEROL, ... | | Authors: | Bingham, C.R, Borhan, B, Geiger, J.H. | | Deposit date: | 2022-07-10 | | Release date: | 2023-02-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | Light controlled reversible Michael addition of cysteine: a new tool for dynamic site-specific labeling of proteins.
Analyst, 148, 2023
|
|
4EDE
 
 | |
4EEJ
 
 | |
4GKC
 
 | |
4ZH6
 
 | |
4ZH9
 
 | |
4ZR2
 
 | |
4I9S
 
 | |
4I9R
 
 | |
1IG7
 
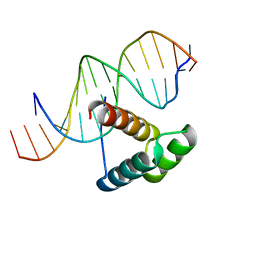 | | Msx-1 Homeodomain/DNA Complex Structure | | Descriptor: | 5'-D(*CP*AP*CP*TP*AP*AP*TP*TP*GP*AP*AP*GP*G)-3', 5'-D(P*TP*CP*CP*TP*TP*CP*AP*AP*TP*TP*AP*GP*TP*GP*AP*C)-3', Homeotic protein Msx-1 | | Authors: | Hovde, S, Abate-Shen, C, Geiger, J.H. | | Deposit date: | 2001-04-17 | | Release date: | 2001-04-23 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of the Msx-1 homeodomain/DNA complex
Biochemistry, 40, 2001
|
|
1KI0
 
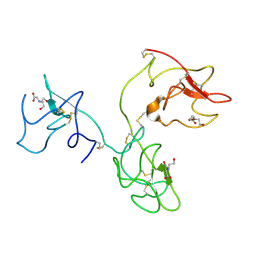 | | The X-ray Structure of Human Angiostatin | | Descriptor: | ANGIOSTATIN, BICINE | | Authors: | Abad, M.C, Arni, R.K, Grella, D.K, Castellino, F.J, Tulinsky, A, Geiger, J.H. | | Deposit date: | 2001-12-02 | | Release date: | 2002-05-29 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The X-ray crystallographic structure of the angiogenesis inhibitor angiostatin.
J.Mol.Biol., 318, 2002
|
|
1M7X
 
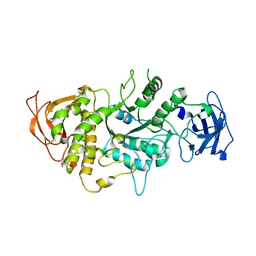 | | The X-ray Crystallographic Structure of Branching Enzyme | | Descriptor: | 1,4-alpha-glucan Branching Enzyme | | Authors: | Abad, M.C, Binderup, K, Rios-Steiner, J, Arni, R.K, Preiss, J, Geiger, J.H. | | Deposit date: | 2002-07-23 | | Release date: | 2002-09-18 | | Last modified: | 2022-12-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The X-ray crystallographic structure of Escherichia coli branching enzyme
J.Biol.Chem., 277, 2002
|
|
4LPC
 
 | |
4M6S
 
 | |
4LQ1
 
 | |
4M7M
 
 | |
3CR6
 
 | |
3F8A
 
 | | Crystal Structure of the R132K:R111L:L121E:R59W Mutant of Cellular Retinoic Acid-Binding Protein Type II Complexed with C15-aldehyde (a retinal analog) at 1.95 Angstrom resolution. | | Descriptor: | 1,3,3-trimethyl-2-[(1E,3E)-3-methylpenta-1,3-dien-1-yl]cyclohexene, 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Cellular retinoic acid-binding protein 2 | | Authors: | Jia, X, Geiger, J.H. | | Deposit date: | 2008-11-12 | | Release date: | 2009-11-10 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Probing Wavelength Regulation with an Engineered Rhodopsin Mimic and a C15-Retinal Analogue
Chempluschem, 77, 2012
|
|
3FA6
 
 | |
3F9D
 
 | |
1YTB
 
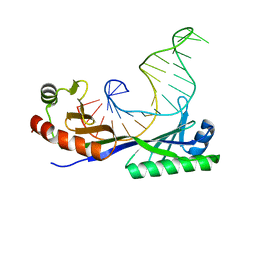 | | CRYSTAL STRUCTURE OF A YEAST TBP/TATA-BOX COMPLEX | | Descriptor: | DNA (29MER), PROTEIN (TATA BINDING PROTEIN (TBP)) | | Authors: | Kim, Y, Geiger, J.H, Hahn, S, Sigler, P.B. | | Deposit date: | 1994-09-28 | | Release date: | 1995-01-26 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of a yeast TBP/TATA-box complex.
Nature, 365, 1993
|
|
1YP2
 
 | | Crystal structure of potato tuber ADP-glucose pyrophosphorylase | | Descriptor: | Glucose-1-phosphate adenylyltransferase small subunit, PARA-MERCURY-BENZENESULFONIC ACID, SULFATE ION | | Authors: | Jin, X, Ballicora, M.A, Preiss, J, Geiger, J.H. | | Deposit date: | 2005-01-29 | | Release date: | 2005-03-15 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Crystal structure of potato tuber ADP-glucose pyrophosphorylase.
Embo J., 24, 2005
|
|
1YP3
 
 | | Crystal structure of potato tuber ADP-glucose pyrophosphorylase in complex with ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Glucose-1-phosphate adenylyltransferase small subunit, SULFATE ION | | Authors: | Jin, X, Ballicora, M.A, Preiss, J, Geiger, J.H. | | Deposit date: | 2005-01-29 | | Release date: | 2005-03-15 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of potato tuber ADP-glucose pyrophosphorylase.
Embo J., 24, 2005
|
|
