6IIC
 
 | | CryoEM structure of Mud Crab Dicistrovirus | | Descriptor: | VP1 of Mud crab dicistrovirus, VP2 of Mud crab dicistrovirus, VP3 of Mud crab dicistrovirus, ... | | Authors: | Zhang, Q, Gao, Y. | | Deposit date: | 2018-10-04 | | Release date: | 2019-01-16 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-electron Microscopy Structures of Novel Viruses from Mud CrabScylla paramamosainwith Multiple Infections.
J. Virol., 93, 2019
|
|
6IZL
 
 | |
6JZZ
 
 | | The crystal structure of AAR-C294S in complex with ADO. | | Descriptor: | Aldehyde decarbonylase, FE (II) ION, HEXADECAN-1-OL, ... | | Authors: | Zhang, H.M, Li, M, Gao, Y. | | Deposit date: | 2019-05-04 | | Release date: | 2020-04-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.011 Å) | | Cite: | Structural insights into catalytic mechanism and product delivery of cyanobacterial acyl-acyl carrier protein reductase.
Nat Commun, 11, 2020
|
|
6JZY
 
 | | Crystal structure of AAR with NADPH and stearyl in complex with ADO binding a long chain carbohydrate | | Descriptor: | Aldehyde decarbonylase, FE (II) ION, HEXADECAN-1-OL, ... | | Authors: | Zhang, H.M, Li, M, Gao, Y. | | Deposit date: | 2019-05-04 | | Release date: | 2020-04-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural insights into catalytic mechanism and product delivery of cyanobacterial acyl-acyl carrier protein reductase.
Nat Commun, 11, 2020
|
|
6JZU
 
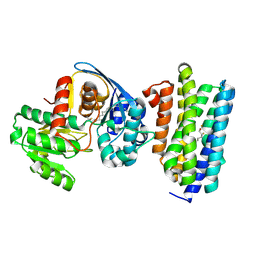 | | The crystal structure of acyl-acyl carrier protein (acyl-ACP) reductase (AAR) in complex with aldehyde deformylating oxygenase (ADO) | | Descriptor: | Aldehyde decarbonylase, FE (II) ION, HEXADECAN-1-OL, ... | | Authors: | Zhang, H.M, Li, M, Gao, Y. | | Deposit date: | 2019-05-03 | | Release date: | 2020-04-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.181 Å) | | Cite: | Structural insights into catalytic mechanism and product delivery of cyanobacterial acyl-acyl carrier protein reductase.
Nat Commun, 11, 2020
|
|
6JZQ
 
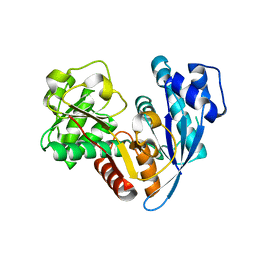 | |
8XGC
 
 | | Structure of yeast replisome associated with FACT and histone hexamer, Composite map | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Cell division control protein 45, Chromosome segregation in meiosis protein 3, ... | | Authors: | Li, N, Gao, Y, Yu, D, Gao, N, Zhai, Y. | | Deposit date: | 2023-12-15 | | Release date: | 2024-02-14 | | Last modified: | 2024-04-10 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Parental histone transfer caught at the replication fork.
Nature, 627, 2024
|
|
4EAY
 
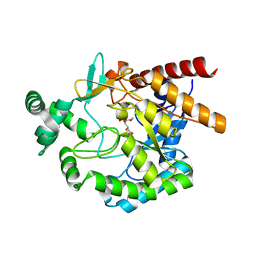 | | Crystal structures of mannonate dehydratase from Escherichia coli strain K12 complexed with D-mannonate | | Descriptor: | CHLORIDE ION, D-MANNONIC ACID, MANGANESE (II) ION, ... | | Authors: | Qiu, X, Zhu, Y, Yuan, Y, Zhang, Y, Liu, H, Gao, Y, Teng, M, Niu, L. | | Deposit date: | 2012-03-23 | | Release date: | 2013-03-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural insights into decreased enzymatic activity induced by an insert sequence in mannonate dehydratase from Gram negative bacterium.
J.Struct.Biol., 180, 2012
|
|
4EAC
 
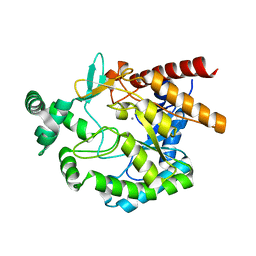 | | Crystal structure of mannonate dehydratase from Escherichia coli strain K12 | | Descriptor: | CHLORIDE ION, MANGANESE (II) ION, Mannonate dehydratase | | Authors: | Qiu, X, Zhu, Y, Yuan, Y, Zhang, Y, Liu, H, Gao, Y, Teng, M, Niu, L. | | Deposit date: | 2012-03-22 | | Release date: | 2013-03-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural insights into decreased enzymatic activity induced by an insert sequence in mannonate dehydratase from Gram negative bacterium.
J.Struct.Biol., 180, 2012
|
|
1J48
 
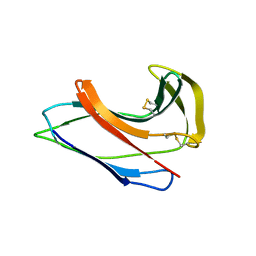 | | Crystal Structure of Apo-C1027 | | Descriptor: | Apoprotein of C1027 | | Authors: | Chen, Y, Li, J, Liu, Y, Bartlam, M, Gao, Y, Jin, L, Tang, H, Shao, Y, Zhen, Y, Rao, Z. | | Deposit date: | 2001-07-30 | | Release date: | 2003-06-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of Apo-C1027 and Computer Modeling Analysis of C1027 Chromophore- Protein Complex
To be published
|
|
7O6E
 
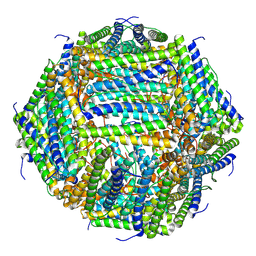 | | 2.12 A cryo-EM structure of Mycobacterium tuberculosis Ferritin | | Descriptor: | Ferritin BfrB | | Authors: | Gijsbers, A, Zhang, Y, Gao, Y, Peters, P.J, Ravelli, R.B.G. | | Deposit date: | 2021-04-10 | | Release date: | 2021-05-19 | | Last modified: | 2021-09-15 | | Method: | ELECTRON MICROSCOPY (2.1 Å) | | Cite: | Mycobacterium tuberculosis ferritin: a suitable workhorse protein for cryo-EM development.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
7P13
 
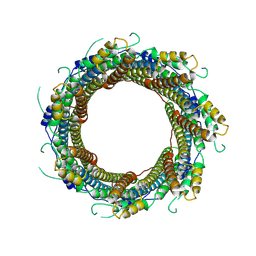 | | 2.29 A Mycobacterium tuberculosis EspB. | | Descriptor: | ESX-1 secretion-associated protein EspB | | Authors: | Gijsbers, A, Zhang, Y, Vinciauskaite, V, Siroy, A, Gao, Y, Tria, G, Mathew, A, Sanchez-Puig, N, Lopez-Iglesias, C, Peters, P.J, Ravelli, R.B.G. | | Deposit date: | 2021-07-01 | | Release date: | 2021-08-18 | | Last modified: | 2021-10-13 | | Method: | ELECTRON MICROSCOPY (2.29 Å) | | Cite: | Priming mycobacterial ESX-secreted protein B to form a channel-like structure.
Curr Res Struct Biol, 3, 2021
|
|
2WLS
 
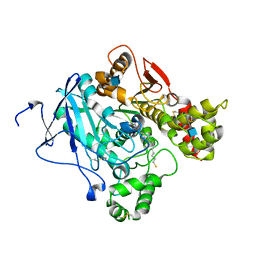 | | Crystal structure of Mus musculus Acetylcholinesterase in complex with AMTS13 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETYLCHOLINESTERASE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Pang, Y.P, Ekstrom, F, Polsinelli, G.A, Gao, Y, Rana, S, Hua, D.H, Andersson, B, Andersson, P.O, Peng, L, Singh, S.K, Mishra, R.K, Zhu, K.Y, Fallon, A.M, Ragsdale, D.W, Brimijoin, S. | | Deposit date: | 2009-06-25 | | Release date: | 2009-09-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Selective and Irreversible Inhibitors of Mosquito Acetylcholinesterases for Controlling Malaria and Other Mosquito-Borne Diseases.
Plos One, 4, 2009
|
|
4YCG
 
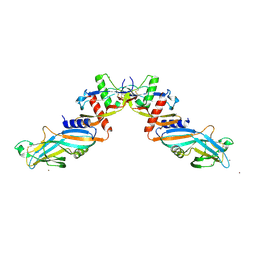 | | Pro-bone morphogenetic protein 9 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Bone Morphogenetic Protein 9 Growth Factor Domain, Bone Morphogenetic Protein 9 Prodomain, ... | | Authors: | Mi, L.-Z, Brown, C.T, Gao, Y, Tian, Y, Le, V, Walz, T, Springer, T.A. | | Deposit date: | 2015-02-20 | | Release date: | 2015-03-04 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structure of bone morphogenetic protein 9 procomplex.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4YCI
 
 | | non-latent pro-bone morphogenetic protein 9 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Bone Morphogenetic Protein 9 Growth Factor Domain, ... | | Authors: | Mi, L.Z, Brown, C.T, Gao, Y, Tian, Y, Le, V, Walz, T, Springer, T.A. | | Deposit date: | 2015-02-20 | | Release date: | 2015-03-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Structure of bone morphogenetic protein 9 procomplex.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
5A9W
 
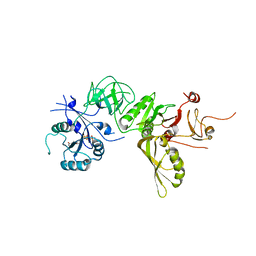 | | Structure of GDPCP BipA | | Descriptor: | GTP-BINDING PROTEIN, PHOSPHOMETHYLPHOSPHONIC ACID GUANYLATE ESTER | | Authors: | Kumar, V, Chen, Y, Ero, R, Li, Z, Gao, Y. | | Deposit date: | 2015-07-23 | | Release date: | 2015-08-26 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Structure of Bipa in GTP Form Bound to the Ratcheted Ribosome.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
5A9V
 
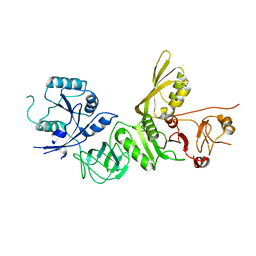 | | Structure of apo BipA | | Descriptor: | GTP-BINDING PROTEIN | | Authors: | Kumar, V, Chen, Y, Ero, R, Li, Z, Gao, Y. | | Deposit date: | 2015-07-23 | | Release date: | 2015-09-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.31 Å) | | Cite: | Structure of Bipa in GTP Form Bound to the Ratcheted Ribosome.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4BHR
 
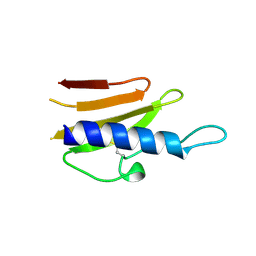 | | Structure of the TTHA1221 type IV pilin protein from Thermus thermophilus | | Descriptor: | PILIN, TYPE IV, SULFATE ION | | Authors: | Karuppiah, V, Collins, R.F, Gao, Y, Thistlethwaite, A, Derrick, J.P. | | Deposit date: | 2013-04-05 | | Release date: | 2013-11-20 | | Last modified: | 2013-12-11 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure and Assembly of an Inner Membrane Platform for Initiation of Type Iv Pilus Biogenesis
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4BHQ
 
 | | Structure of the periplasmic domain of the PilN type IV pilus biogenesis protein from Thermus thermophilus | | Descriptor: | COMPETENCE PROTEIN PILN, MAGNESIUM ION | | Authors: | Karuppiah, V, Collins, R.F, Gao, Y, Thistlethwaite, A, Derrick, J.P. | | Deposit date: | 2013-04-05 | | Release date: | 2013-11-20 | | Last modified: | 2013-12-11 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structure and Assembly of an Inner Membrane Platform for Initiation of Type Iv Pilus Biogenesis
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4DM4
 
 | | The conserved domain of yeast Cdc73 | | Descriptor: | Cell division control protein 73 | | Authors: | Chen, H, Shi, N, Gao, Y, Li, X, Niu, L, Teng, M. | | Deposit date: | 2012-02-06 | | Release date: | 2012-08-22 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Crystallographic analysis of the conserved C-terminal domain of transcription factor Cdc73 from Saccharomyces cerevisiae reveals a GTPase-like fold.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
2OGD
 
 | | T. Brucei Farnesyl Diphosphate Synthase Complexed with Bisphosphonate BPH-527 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ACETATE ION, BETA-MERCAPTOETHANOL, ... | | Authors: | Cao, R, Gao, Y, Robinson, H, Goddard, A, Oldfield, E. | | Deposit date: | 2007-01-05 | | Release date: | 2007-10-02 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Bisphosphonates: Teaching Old Drugs with New Tricks
TO BE PUBLISHED
|
|
6O8A
 
 | | Thaumatin native-SAD structure determined at 5 keV from microcrystals | | Descriptor: | L(+)-TARTARIC ACID, Thaumatin-1 | | Authors: | Guo, G, Zhu, P, Fuchs, M.R, Shi, W, Andi, B, Gao, Y, Hendrickson, W.A, McSweeney, S, Liu, Q. | | Deposit date: | 2019-03-09 | | Release date: | 2019-05-08 | | Last modified: | 2019-12-18 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Synchrotron microcrystal native-SAD phasing at a low energy.
Iucrj, 6, 2019
|
|
1K5W
 
 | | THREE-DIMENSIONAL STRUCTURE OF THE SYNAPTOTAGMIN 1 C2B-DOMAIN: SYNAPTOTAGMIN 1 AS A PHOSPHOLIPID BINDING MACHINE | | Descriptor: | CALCIUM ION, Synaptotagmin I | | Authors: | Fernandez, I, Arac, D, Ubach, J, Gerber, S.H, Shin, O, Gao, Y, Anderson, R.G.W, Sudhof, T.C, Rizo, J. | | Deposit date: | 2001-10-12 | | Release date: | 2002-01-23 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional structure of the synaptotagmin 1 C2B-domain: synaptotagmin 1 as a phospholipid binding machine.
Neuron, 32, 2001
|
|
8EF9
 
 | | Structure of Lates calcarifer DNA polymerase theta polymerase domain with long duplex DNA, complex Ia | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, DNA (5'-D(*AP*GP*CP*AP*TP*CP*CP*GP*TP*AP*GP*(2DA))-3'), DNA (5'-D(*AP*GP*CP*TP*CP*TP*AP*CP*GP*GP*AP*TP*GP*C)-3'), ... | | Authors: | Li, C, Zhu, H, Sun, J, Gao, Y. | | Deposit date: | 2022-09-08 | | Release date: | 2022-12-14 | | Last modified: | 2023-02-01 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | Structural basis of DNA polymerase theta mediated DNA end joining.
Nucleic Acids Res., 51, 2023
|
|
8EFC
 
 | | Structure of Lates calcarifer DNA polymerase theta polymerase domain with long duplex DNA, complex Ia | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, DNA (5'-D(*AP*CP*TP*GP*TP*GP*AP*GP*GP*CP*AP*TP*CP*CP*GP*TP*AP*GP*(2DA))-3'), DNA (5'-D(*AP*GP*CP*TP*CP*TP*AP*CP*GP*GP*AP*TP*GP*CP*CP*TP*CP*AP*CP*AP*G)-3'), ... | | Authors: | Li, C, Zhu, H, Sun, J, Gao, Y. | | Deposit date: | 2022-09-08 | | Release date: | 2022-12-14 | | Last modified: | 2023-02-01 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural basis of DNA polymerase theta mediated DNA end joining.
Nucleic Acids Res., 51, 2023
|
|
