4MOD
 
 | | Structure of the MERS-CoV fusion core | | Descriptor: | HR1 of S protein, LINKER, HR2 of S protein | | Authors: | Gao, J, Lu, G, Qi, J, Li, Y, Wu, Y, Deng, Y, Geng, H, Xiao, H, Tan, W, Yan, J, Gao, G.F. | | Deposit date: | 2013-09-12 | | Release date: | 2013-10-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.901 Å) | | Cite: | Structure of the fusion core and inhibition of fusion by a heptad repeat peptide derived from the S protein of Middle East respiratory syndrome coronavirus.
J.Virol., 87, 2013
|
|
1BO9
 
 | |
8DL5
 
 | |
4KS7
 
 | | PAK6 kinase domain in complex with PF-3758309 | | Descriptor: | ISOPROPYL ALCOHOL, PF-3758309, Serine/threonine-protein kinase PAK 6 | | Authors: | Gao, J, Boggon, T.J. | | Deposit date: | 2013-05-17 | | Release date: | 2013-09-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Substrate and Inhibitor Specificity of the Type II p21-Activated Kinase, PAK6.
Plos One, 8, 2013
|
|
4KS8
 
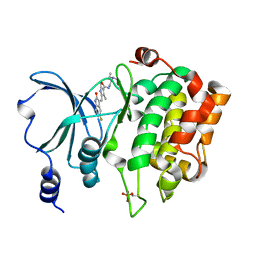 | | PAK6 kinase domain in complex with sunitinib | | Descriptor: | N-[2-(diethylamino)ethyl]-5-[(Z)-(5-fluoro-2-oxo-1,2-dihydro-3H-indol-3-ylidene)methyl]-2,4-dimethyl-1H-pyrrole-3-carbo xamide, Serine/threonine-protein kinase PAK 6 | | Authors: | Gao, J, Boggon, T.J. | | Deposit date: | 2013-05-17 | | Release date: | 2013-09-11 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Substrate and Inhibitor Specificity of the Type II p21-Activated Kinase, PAK6.
Plos One, 8, 2013
|
|
5GLY
 
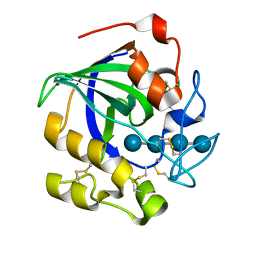 | | Crystal structure of a glycoside hydrolase in complex with cellotetrose from Thielavia terrestris NRRL 8126 | | Descriptor: | Glycoside hydrolase family 45 protein, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-alpha-D-glucopyranose, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Gao, J, Liu, W.D, Zheng, Y.Y, Chen, C.C, Guo, R.T. | | Deposit date: | 2016-07-12 | | Release date: | 2017-04-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Characterization and crystal structure of a thermostable glycoside hydrolase family 45 1,4-beta-endoglucanase from Thielavia terrestris
Enzyme Microb. Technol., 99, 2017
|
|
5GLX
 
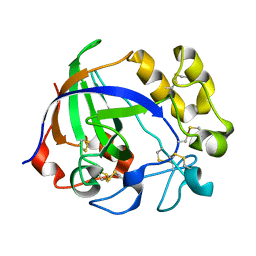 | | Crystal structure of a glycoside hydrolase from Thielavia terrestris NRRL 8126 | | Descriptor: | Glycoside hydrolase family 45 protein | | Authors: | Gao, J, Liu, W.D, Zheng, Y.Y, Chen, C.C, Guo, R.T. | | Deposit date: | 2016-07-12 | | Release date: | 2017-04-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Characterization and crystal structure of a thermostable glycoside hydrolase family 45 1,4-beta-endoglucanase from Thielavia terrestris
Enzyme Microb. Technol., 99, 2017
|
|
5GM9
 
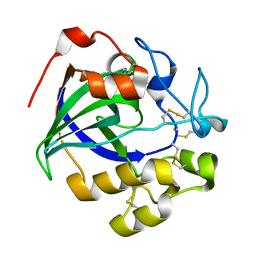 | | Crystal structure of a glycoside hydrolase in complex with cellobiose | | Descriptor: | Glycoside hydrolase family 45 protein, beta-D-glucopyranose-(1-4)-alpha-D-glucopyranose, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Gao, J, Liu, W.D, Zheng, Y.Y, Chen, C.C, Guo, R.T. | | Deposit date: | 2016-07-13 | | Release date: | 2017-04-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Characterization and crystal structure of a thermostable glycoside hydrolase family 45 1,4-beta-endoglucanase from Thielavia terrestris
Enzyme Microb. Technol., 99, 2017
|
|
8WEX
 
 | |
6IF4
 
 | | Crystal structure of Tbtudor | | Descriptor: | Histone acetyltransferase | | Authors: | Gao, J, Ye, K, Diwu, Y, Liao, S, Tu, X. | | Deposit date: | 2018-09-18 | | Release date: | 2019-09-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.934 Å) | | Cite: | Crystal structure of TbEsa1 presumed Tudor domain from Trypanosoma brucei.
J.Struct.Biol., 209, 2020
|
|
8IJT
 
 | | crystal structure of Hyp N135A mutant from Hypoxylon sp. E7406B | | Descriptor: | Terpene synthase | | Authors: | Gao, J, Su, L.Q, Li, Q, Han, X, Wei, H.L, Dai, Z.J, Liu, W.D. | | Deposit date: | 2023-02-28 | | Release date: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | crystal structure of Hyp N135A mutant from Hypoxylon sp. E7406B
to be published
|
|
8II9
 
 | | crystal structure of Hyp mutant from Hypoxylon sp. E7406B | | Descriptor: | Terpene synthase | | Authors: | Gao, J, Liu, W.D, Li, Q, Han, X, Wei, H.L, Dai, Z.J, Su, L.Q. | | Deposit date: | 2023-02-24 | | Release date: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | crystal structure of Hyp
to be published
|
|
7CNF
 
 | | Solution strcture of HsTFIIS LW domain | | Descriptor: | Transcription elongation factor A protein 1 | | Authors: | Gao, J, Liao, S. | | Deposit date: | 2020-07-31 | | Release date: | 2021-08-04 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution strcture of HsTFIIS LW domain
To Be Published
|
|
7K37
 
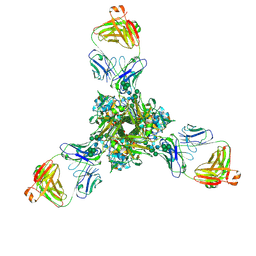 | | Structure of full-length influenza HA with a head-binding antibody at pH 7.8 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin, ... | | Authors: | Gui, M, Xiang, Y, Gao, J. | | Deposit date: | 2020-09-10 | | Release date: | 2020-11-11 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural intermediates in the low pH-induced transition of influenza hemagglutinin.
Plos Pathog., 16, 2020
|
|
7K39
 
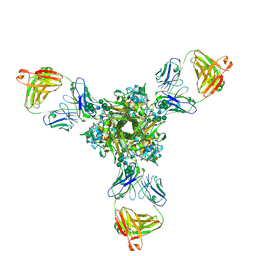 | | Structure of full-length influenza HA with a head-binding antibody at pH 5.2, conformation A, neutral pH-like | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin, ... | | Authors: | Gui, M, Gao, J, Xiang, Y. | | Deposit date: | 2020-09-10 | | Release date: | 2020-11-11 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural intermediates in the low pH-induced transition of influenza hemagglutinin.
Plos Pathog., 16, 2020
|
|
7K3B
 
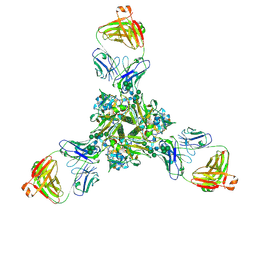 | | Structure of full-length influenza HA with a head-binding antibody at pH 5.2, conformation C, central helices splay | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin, ... | | Authors: | Gui, M, Gao, J, Xiang, Y. | | Deposit date: | 2020-09-10 | | Release date: | 2020-11-11 | | Last modified: | 2020-12-09 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural intermediates in the low pH-induced transition of influenza hemagglutinin.
Plos Pathog., 16, 2020
|
|
7K3A
 
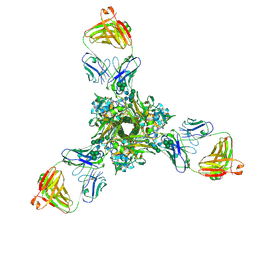 | | Structure of full-length influenza HA with a head-binding antibody at pH 5.2, conformation B, fusion peptide release | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin, ... | | Authors: | Gui, M, Gao, J, Xiang, Y. | | Deposit date: | 2020-09-10 | | Release date: | 2020-11-11 | | Last modified: | 2020-12-09 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Structural intermediates in the low pH-induced transition of influenza hemagglutinin.
Plos Pathog., 16, 2020
|
|
9AZB
 
 | | Crystal structure of LolTv5 | | Descriptor: | Aminotransferase, class V/Cysteine desulfurase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Gao, J, Hai, Y. | | Deposit date: | 2024-03-11 | | Release date: | 2024-07-17 | | Last modified: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Enzymatic Synthesis of Unprotected alpha , beta-Diamino Acids via Direct Asymmetric Mannich Reactions.
J.Am.Chem.Soc., 146, 2024
|
|
9AZA
 
 | | Crystal structure of LolTv4 | | Descriptor: | Aminotransferase, class V/Cysteine desulfurase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Gao, J, Hai, Y. | | Deposit date: | 2024-03-10 | | Release date: | 2024-07-17 | | Last modified: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | Enzymatic Synthesis of Unprotected alpha , beta-Diamino Acids via Direct Asymmetric Mannich Reactions.
J.Am.Chem.Soc., 146, 2024
|
|
7XGW
 
 | |
7YJI
 
 | |
3WYJ
 
 | | Structure of E. coli undecaprenyl diphosphate synthase in complex with BPH-789 | | Descriptor: | Ditrans,polycis-undecaprenyl-diphosphate synthase ((2E,6E)-farnesyl-diphosphate specific), [1-oxidanyl-2-[3-[3-[[3-[[3-[3-(2-oxidanyl-2,2-diphosphono-ethyl)phenyl]phenyl]sulfamoyl]phenyl]sulfonylamino]phenyl]phenyl]-1-phosphono-ethyl]phosphonic acid | | Authors: | Gao, J, Ko, T.P, Huang, C.H, Oldfield, E, Guo, R.T. | | Deposit date: | 2014-08-29 | | Release date: | 2015-02-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Antibacterial drug leads: DNA and enzyme multitargeting.
J.Med.Chem., 58, 2015
|
|
3WYI
 
 | | Structure of S. aureus undecaprenyl diphosphate synthase | | Descriptor: | Isoprenyl transferase, MAGNESIUM ION | | Authors: | Gao, J, Ko, T.P, Huang, C.H, Oldfield, E, Guo, R.T. | | Deposit date: | 2014-08-29 | | Release date: | 2015-02-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Antibacterial drug leads: DNA and enzyme multitargeting.
J.Med.Chem., 58, 2015
|
|
7W66
 
 | | Crystal structure of a PSH1 mutant in complex with ligand | | Descriptor: | PSH1, bis(2-hydroxyethyl) benzene-1,4-dicarboxylate | | Authors: | Gao, J, Lara, P, Li, Z.S, Han, X, Wei, R, Liu, W.D. | | Deposit date: | 2021-12-01 | | Release date: | 2022-09-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Multiple Substrate Binding Mode-Guided Engineering of a Thermophilic PET Hydrolase.
Acs Catalysis, 12, 2022
|
|
7W6C
 
 | | Crystal structure of a PSH1 in complex with ligand J1K | | Descriptor: | 4-(2-hydroxyethylcarbamoyl)benzoic acid, PSH1 | | Authors: | Gao, J, Lara, P, Li, Z.S, Han, X, Wei, R, Liu, W.D. | | Deposit date: | 2021-12-01 | | Release date: | 2022-09-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Multiple Substrate Binding Mode-Guided Engineering of a Thermophilic PET Hydrolase.
Acs Catalysis, 12, 2022
|
|
