6O9B
 
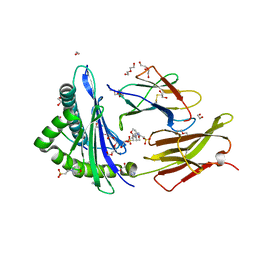 | |
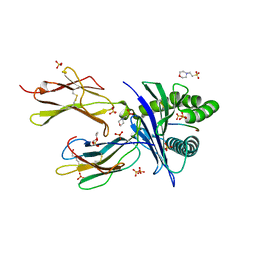
6O9C
 
 | |
1MR2
 
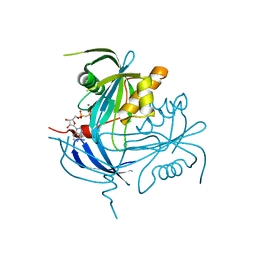 | | Structure of the MT-ADPRase in complex with 1 Mn2+ ion and AMP-CP (a inhibitor), a nudix enzyme | | Descriptor: | ADPR pyrophosphatase, MANGANESE (II) ION, PHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER | | Authors: | Kang, L.-W, Gabelli, S.B, Bianchet, M.A, Cunningham, J.E, O'Handley, S.F, Amzel, L.M. | | Deposit date: | 2002-09-17 | | Release date: | 2003-08-05 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure and mechanism of MT-ADPRase, a Nudix hydrolase from Mycobacterium tuberculosis
Structure, 11, 2003
|
|
1MQW
 
 | | Structure of the MT-ADPRase in complex with three Mn2+ ions and AMPCPR, a Nudix enzyme | | Descriptor: | ADPR pyrophosphatase, ALPHA-BETA METHYLENE ADP-RIBOSE, MANGANESE (II) ION | | Authors: | Kang, L.-W, Gabelli, S.B, Bianchet, M.A, Cunningham, J.E, O'Handley, S.F, Amzel, L.M. | | Deposit date: | 2002-09-17 | | Release date: | 2003-08-05 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure and mechanism of MT-ADPRase, a Nudix hydrolase from Mycobacterium tuberculosis
Structure, 11, 2003
|
|
1N8Z
 
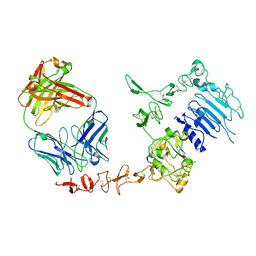 | | Crystal structure of extracellular domain of human HER2 complexed with Herceptin Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Herceptin Fab (antibody) - heavy chain, Herceptin Fab (antibody) - light chain, ... | | Authors: | Cho, H.-S, Mason, K, Ramyar, K.X, Stanley, A.M, Gabelli, S.B, Denney Jr, D.W, Leahy, D.J. | | Deposit date: | 2002-11-21 | | Release date: | 2003-02-18 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Structure of the Extracellular Region of HER2 Alone and in Complex with the Herceptin Fab
Nature, 421, 2003
|
|
1N8Y
 
 | | Crystal structure of the extracellular region of rat HER2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, protooncoprotein | | Authors: | Cho, H.-S, Mason, K, Ramyar, K.X, Stanley, A.M, Gabelli, S.B, Denney Jr, D.W, Leahy, D.J. | | Deposit date: | 2002-11-21 | | Release date: | 2003-02-18 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of the Extracellular Region of HER2 Alone and in complex with the Herceptin Fab
Nature, 421, 2003
|
|
1NQZ
 
 | | The structure of a CoA pyrophosphatase from D. Radiodurans complexed with a magnesium ion | | Descriptor: | CoA pyrophosphatase (MutT/nudix family protein), MAGNESIUM ION | | Authors: | Kang, L.W, Gabelli, S.B, Bianchet, M.A, Xu, W.L, Bessman, M.J, Amzel, L.M. | | Deposit date: | 2003-01-23 | | Release date: | 2003-05-13 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of a coenzyme A pyrophosphatase from Deinococcus radiodurans: a member of the Nudix family.
J.Bacteriol., 185, 2003
|
|
1NQY
 
 | | The structure of a CoA pyrophosphatase from D. Radiodurans | | Descriptor: | CoA pyrophosphatase (MutT/nudix family protein) | | Authors: | Kang, L.W, Gabelli, S.B, Bianchet, M.A, Xu, W.L, Bessman, M.J, Amzel, L.M. | | Deposit date: | 2003-01-23 | | Release date: | 2003-05-13 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Structure of a coenzyme A pyrophosphatase from Deinococcus radiodurans: a member of the Nudix family.
J.Bacteriol., 185, 2003
|
|
7JUK
 
 | | Crystal structure of PTEN with a tetra-phosphorylated tail (4p-crPTEN-13sp-T2, SDTTDSDPENEG) | | Descriptor: | PHOSPHATE ION, Phosphatidylinositol 3,4,5-trisphosphate 3-phosphatase and dual-specificity protein phosphatase PTEN | | Authors: | Dempsey, D, Phan, K, Cole, P, Gabelli, S.B. | | Deposit date: | 2020-08-19 | | Release date: | 2021-10-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | The structural basis of PTEN regulation by multi-site phosphorylation.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7KGU
 
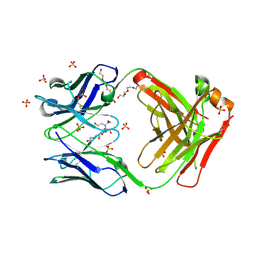 | | Structure of 2Q1-Fab, an antibody selective for IDH2R140Q-HLA-B*07:02 | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Miller, M.S, Aytenfisu, T.Y, Wright, K.M, Gabelli, S.B. | | Deposit date: | 2020-10-18 | | Release date: | 2021-06-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural engineering of chimeric antigen receptors targeting HLA-restricted neoantigens.
Nat Commun, 12, 2021
|
|
7JVX
 
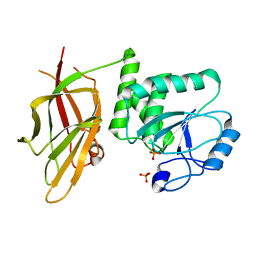 | | Crystal structure of PTEN (aa 7-353 followed by spacer TGGGSGGTGGGSGGTGGGCY ligated to peptide pSDpTpTDpSDPENEPFDED) | | Descriptor: | PHOSPHATE ION, Phosphatidylinositol 3,4,5-trisphosphate 3-phosphatase and dual-specificity protein phosphatase PTEN | | Authors: | Dempsey, D, Phan, K, Cole, P, Gabelli, S.B. | | Deposit date: | 2020-08-24 | | Release date: | 2021-08-04 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The structural basis of PTEN regulation by multi-site phosphorylation.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7KU0
 
 | | Data clustering and dynamics of chymotrypsinogen cluster 138 (yellow) structure | | Descriptor: | Chymotrypsinogen A, SULFATE ION | | Authors: | Nguyen, T, Phan, K.L, Kreitler, D.F, Andrews, L.C, Gabelli, S.B, Kozakov, D, Jakoncic, J, Sweet, R.M, Soares, A.S, Bernstein, H.J. | | Deposit date: | 2020-11-24 | | Release date: | 2021-03-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | A simple technique to classify diffraction data from dynamic proteins according to individual polymorphs.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7KTY
 
 | | Data clustering and dynamics of chymotrypsinogen average structure | | Descriptor: | Chymotrypsinogen A, SULFATE ION | | Authors: | Nguyen, T, Phan, K.L, Kreitler, D.F, Andrews, L.C, Gabelli, S.B, Kozakov, D, Jakoncic, J, Shi, W, Sweet, R.M, Soares, A.S, Bernstein, H.J. | | Deposit date: | 2020-11-24 | | Release date: | 2021-03-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A simple technique to classify diffraction data from dynamic proteins according to individual polymorphs.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7KU1
 
 | | Data clustering and dynamics of chymotrypsinogen cluster 139 (green) structure | | Descriptor: | Chymotrypsinogen A, SULFATE ION | | Authors: | Nguyen, T, Phan, K.L, Kreitler, D.F, Andrews, L.C, Gabelli, S.B, Kozakov, D, Jakoncic, J, Sweet, R.M, Soares, A.S, Bernstein, H.J. | | Deposit date: | 2020-11-24 | | Release date: | 2021-03-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | A simple technique to classify diffraction data from dynamic proteins according to individual polymorphs.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7KU2
 
 | | Data clustering and dynamics of chymotrypsinogen clulster 140 (structure) | | Descriptor: | Chymotrypsinogen A, SULFATE ION | | Authors: | Nguyen, T, Phan, K.L, Kreitler, D.F, Andrews, L.C, Gabelli, S.B, Kozakov, D, Jakoncic, J, Sweet, R.M, Soares, A.S, Bernstein, H.J. | | Deposit date: | 2020-11-24 | | Release date: | 2021-03-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.185 Å) | | Cite: | A simple technique to classify diffraction data from dynamic proteins according to individual polymorphs.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7KU3
 
 | | Data clustering and dynamics of chymotrypsinogen cluster 141 (cyan) structure | | Descriptor: | Chymotrypsinogen A, SULFATE ION | | Authors: | Nguyen, T, Phan, K.L, Kreitler, D.F, Andrews, L.C, Gabelli, S.B, Kozakov, D, Jakoncic, J, Sweet, R.M, Soares, A.S, Bernstein, H.J. | | Deposit date: | 2020-11-24 | | Release date: | 2021-03-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A simple technique to classify diffraction data from dynamic proteins according to individual polymorphs.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7KTZ
 
 | | Data clustering and dynamics of chymotrypsinogen cluster 131 (purple) structure | | Descriptor: | Chymotrypsinogen A, SULFATE ION | | Authors: | Nguyen, T, Phan, K.L, Kreitler, D.F, Andrews, L.C, Gabelli, S.B, Kozakov, D, Jakoncic, J, Sweet, R.M, Soares, A.S, Bernstein, H.J. | | Deposit date: | 2020-11-24 | | Release date: | 2021-03-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A simple technique to classify diffraction data from dynamic proteins according to individual polymorphs.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
6ALA
 
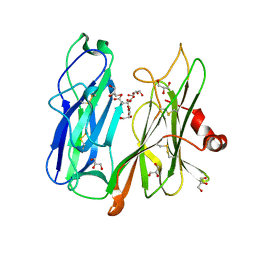 | | Crystal structure of H108A peptidylglycine alpha-hydroxylating monooxygenase (PHM) in complex with citrate | | Descriptor: | CITRATE ANION, COPPER (II) ION, GLYCEROL, ... | | Authors: | Maheshwari, S, Rudzka, K, Gabelli, S.B, Amzel, L.M. | | Deposit date: | 2017-08-07 | | Release date: | 2018-07-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Effects of copper occupancy on the conformational landscape of peptidylglycine alpha-hydroxylating monooxygenase.
Commun Biol, 1, 2018
|
|
6ALV
 
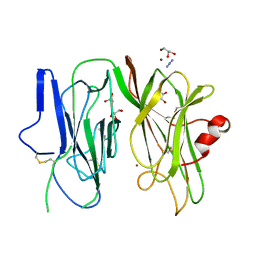 | | Crystal structure of H107A-peptidylglycine alpha-hydroxylating monooxygenase (PHM) mutant (no CuH bound) | | Descriptor: | AZIDE ION, COPPER (II) ION, GLYCEROL, ... | | Authors: | Maheshwari, S, Rudzka, K, Gabelli, S.B, Amzel, L.M. | | Deposit date: | 2017-08-08 | | Release date: | 2018-07-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Effects of copper occupancy on the conformational landscape of peptidylglycine alpha-hydroxylating monooxygenase.
Commun Biol, 1, 2018
|
|
6AN3
 
 | | Crystal structure of H172A-peptidylglycine alpha-hydroxylating monooxygenase (PHM) mutant soaked with peptide (no CuH bound, no peptide bound) | | Descriptor: | COPPER (II) ION, DI(HYDROXYETHYL)ETHER, Peptidyl-glycine alpha-amidating monooxygenase | | Authors: | Maheshwari, S, Rudzka, K, Gabelli, S.B, Amzel, L.M. | | Deposit date: | 2017-08-11 | | Release date: | 2018-07-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Effects of copper occupancy on the conformational landscape of peptidylglycine alpha-hydroxylating monooxygenase.
Commun Biol, 1, 2018
|
|
6AY0
 
 | | Crystal structure of H108A peptidylglycine alpha-hydroxylating monooxygenase (PHM) soaked with peptide | | Descriptor: | COPPER (II) ION, Peptidyl-glycine alpha-amidating monooxygenase | | Authors: | Maheshwari, S, Rudzka, K, Gabelli, S.B, Amzel, L.M. | | Deposit date: | 2017-09-07 | | Release date: | 2018-07-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Effects of copper occupancy on the conformational landscape of peptidylglycine alpha-hydroxylating monooxygenase.
Commun Biol, 1, 2018
|
|
6AMP
 
 | | Crystal structure of H172A PHM (CuH absent, CuM present) | | Descriptor: | COPPER (II) ION, Peptidyl-glycine alpha-amidating monooxygenase | | Authors: | Maheshwari, S, Rudzka, K, Gabelli, S.B, Amzel, L.M. | | Deposit date: | 2017-08-10 | | Release date: | 2018-07-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Effects of copper occupancy on the conformational landscape of peptidylglycine alpha-hydroxylating monooxygenase.
Commun Biol, 1, 2018
|
|
6AO6
 
 | | Crystal structure of H108A peptidylglycine alpha-hydroxylating monooxygenase (PHM) | | Descriptor: | COPPER (II) ION, GLYCEROL, NICKEL (II) ION, ... | | Authors: | Maheshwari, S, Rudzka, K, Gabelli, S.B, Amzel, L.M. | | Deposit date: | 2017-08-15 | | Release date: | 2018-07-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.98 Å) | | Cite: | Effects of copper occupancy on the conformational landscape of peptidylglycine alpha-hydroxylating monooxygenase.
Commun Biol, 1, 2018
|
|
6B09
 
 | |
3NPE
 
 | | Structure of VP14 in complex with oxygen | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, 9-cis-epoxycarotenoid dioxygenase 1, chloroplastic, ... | | Authors: | Messing, S.A, Gabelli, S.B, Amzel, L.M. | | Deposit date: | 2010-06-28 | | Release date: | 2010-11-10 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural insights into maize viviparous14, a key enzyme in the biosynthesis of the phytohormone abscisic acid.
Plant Cell, 22, 2010
|
|
