1BAK
 
 | |
2MWS
 
 | |
8E7O
 
 | | CRYSTAL STRUCTURE OF LYS48-LINKED TETRAUBIQUITIN | | Descriptor: | SULFATE ION, Ubiquitin | | Authors: | Lemma, B.E, Fushman, D. | | Deposit date: | 2022-08-24 | | Release date: | 2022-11-02 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Mechanism of selective recognition of Lys48-linked polyubiquitin by macrocyclic peptide inhibitors of proteasomal degradation.
Nat Commun, 14, 2023
|
|
8F1F
 
 | | Structure of K48-linked tri-ubiquitin in complex with cyclic peptide | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, GLYCEROL, Non-proteinogenic cyclic peptide (inhibitor), ... | | Authors: | Lubkowski, J, Fushman, D, Lemma, B. | | Deposit date: | 2022-11-05 | | Release date: | 2023-11-01 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Mechanism of selective recognition of Lys48-linked polyubiquitin by macrocyclic peptide inhibitors of proteasomal degradation.
Nat Commun, 14, 2023
|
|
2BGF
 
 | |
1XBH
 
 | | A BETA-HAIRPIN MIMIC FROM FCERI-ALPHA-CYCLO(L-262) | | Descriptor: | PROTEIN (CYCLO(L-262)) | | Authors: | Mcdonnell, J.M, Fushman, D, Cahill, S.M, Sutton, B.J, Cowburn, D. | | Deposit date: | 1999-02-17 | | Release date: | 1999-02-21 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Solution Structures of FceRI Alpha-Chain Mimics: A Beta-Hairpin Peptide and Its Retroenantiomer
J.Am.Chem.Soc., 119, 1997
|
|
2PE9
 
 | |
2PEA
 
 | |
3NS8
 
 | |
1M30
 
 | | Solution structure of N-terminal SH3 domain from oncogene protein c-Crk | | Descriptor: | Proto-oncogene C-crk | | Authors: | Schumann, F.H, Varadan, R, Tayakuniyil, P.P, Hall, J.B, Camarero, J.A, Fushman, D. | | Deposit date: | 2002-06-26 | | Release date: | 2003-08-05 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Changing protein backbone topology: Structural and dynamic consequences of the backbone cyclization in SH3 domain
To be Published
|
|
1M3C
 
 | | Solution structure of a circular form of the N-terminal SH3 domain (E132C, E133G, R191G mutant) from oncogene protein c-Crk | | Descriptor: | Proto-oncogene C-crk | | Authors: | Schumann, F.H, Varadan, R, Tayakuniyil, P.P, Hall, J.B, Camarero, J.A, Fushman, D. | | Deposit date: | 2002-06-27 | | Release date: | 2003-08-05 | | Last modified: | 2021-10-27 | | Method: | SOLUTION NMR | | Cite: | Changing protein backbone topology: Structural and dynamic consequences of the backbone cyclization in SH3 domain
To be Published
|
|
5EMZ
 
 | |
1M3B
 
 | | Solution structure of a circular form of the N-terminal SH3 domain (A134C, E135G, R191G mutant) from oncogene protein c-Crk. | | Descriptor: | Proto-oncogene C-crk | | Authors: | Schumann, F.H, Varadan, R, Tayakuniyil, P.P, Hall, J.B, Camarero, J.A, Fushman, D. | | Deposit date: | 2002-06-27 | | Release date: | 2003-08-05 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Changing protein backbone topology: Structural and dynamic consequences of the backbone cyclization in SH3 domain
To be Published
|
|
1M3A
 
 | | Solution structure of a circular form of the truncated N-terminal SH3 domain from oncogene protein c-Crk. | | Descriptor: | Proto-oncogene C-crk | | Authors: | Schumann, F.H, Varadan, R, Tayakuniyil, P.P, Hall, J.B, Camarero, J.A, Fushman, D. | | Deposit date: | 2002-06-27 | | Release date: | 2003-08-05 | | Last modified: | 2024-10-09 | | Method: | SOLUTION NMR | | Cite: | Changing protein backbone topology: Structural and dynamic consequences of the backbone cyclization in SH3 domain
To be Published
|
|
7RBR
 
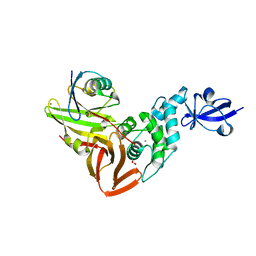 | | The crystal structure of Papain-Like Protease of SARS CoV-2, C111S mutant, in complex with a Lys48-linked di-ubiquitin | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Papain-like protease, ... | | Authors: | Osipiuk, J, Tesar, C, Endres, M, Lanham, B.T, Wydorski, P, Fushman, D, Joachimiak, L, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2021-07-06 | | Release date: | 2021-09-29 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Dual domain recognition determines SARS-CoV-2 PLpro selectivity for human ISG15 and K48-linked di-ubiquitin.
Nat Commun, 14, 2023
|
|
7UV5
 
 | | The crystal structure of Papain-Like Protease of SARS CoV-2, C111S/D286N mutant, in complex with a Lys48-linked di-ubiquitin | | Descriptor: | 1,2-ETHANEDIOL, Papain-like protease nsp3, Ubiquitin, ... | | Authors: | Osipiuk, J, Tesar, C, Endres, M, Lanham, B.T, Wydorski, P, Fushman, D, Joachimiak, L, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2022-04-29 | | Release date: | 2022-05-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Dual domain recognition determines SARS-CoV-2 PLpro selectivity for human ISG15 and K48-linked di-ubiquitin.
Nat Commun, 14, 2023
|
|
1DDB
 
 | | STRUCTURE OF MOUSE BID, NMR, 20 STRUCTURES | | Descriptor: | PROTEIN (BID) | | Authors: | Mcdonnell, J.M, Fushman, D, Milliman, C, Korsmeyer, S.J, Cowburn, D. | | Deposit date: | 1999-02-19 | | Release date: | 1999-08-30 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the proapoptotic molecule BID: a structural basis for apoptotic agonists and antagonists.
Cell(Cambridge,Mass.), 96, 1999
|
|
5UJL
 
 | |
5UJN
 
 | |
6OQ1
 
 | |
6OQ2
 
 | |
7S6O
 
 | | The crystal structure of Lys48-linked di-ubiquitin | | Descriptor: | ACETATE ION, Ubiquitin | | Authors: | Osipiuk, J, Tesar, C, Lanham, B.T, Wydorski, P, Fushman, D, Joachimiak, L, Joachimiak, A. | | Deposit date: | 2021-09-14 | | Release date: | 2021-09-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Dual domain recognition determines SARS-CoV-2 PLpro selectivity for human ISG15 and K48-linked di-ubiquitin.
Nat Commun, 14, 2023
|
|
1C4B
 
 | | A BETA-HAIRPIN MIMIC FROM FCERI-ALPHA-CYCLO(RD-262) | | Descriptor: | PROTEIN (CYCLO(RD-262)) | | Authors: | Mcdonnell, J.M, Fushman, D, Cahill, S.M, Sutton, B.J, Cowburn, D. | | Deposit date: | 1999-08-02 | | Release date: | 1999-08-25 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution Structures of FceRI Alpha-Chain Mimics: A Beta-Hairpin Peptide and Its Retroenantiomer
J.Am.Chem.Soc., 119, 1997
|
|
1A6S
 
 | | M-DOMAIN FROM GAG POLYPROTEIN OF ROUS SARCOMA VIRUS, NMR, 20 STRUCTURES | | Descriptor: | GAG POLYPROTEIN | | Authors: | Mcdonnell, J.M, Fushman, D, Cahill, S.M, Zhou, W, Wolven, A, Wilson, C.B, Nelle, T.D, Resh, M.D, Wills, J, Cowburn, D. | | Deposit date: | 1998-03-02 | | Release date: | 1998-10-14 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure and dynamics of the bioactive retroviral M domain from Rous sarcoma virus
J.Mol.Biol., 279, 1998
|
|
5W46
 
 | |
