8T1Y
 
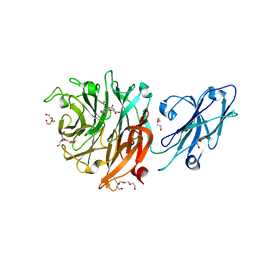 | |
8T1Z
 
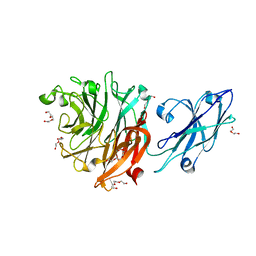 | |
8T26
 
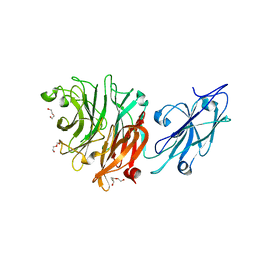 | |
8T24
 
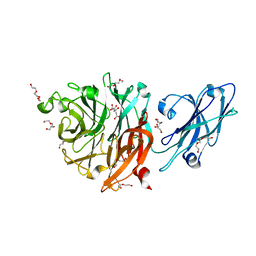 | |
8T27
 
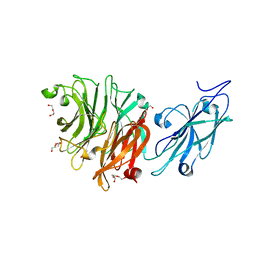 | |
8FEB
 
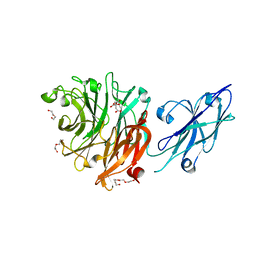 | | Crystal Structure of Porphyromonas gingivalis Sialidase (PG_0352) | | Descriptor: | CITRATE ANION, DI(HYDROXYETHYL)ETHER, Sialidase, ... | | Authors: | Clark, N.D, Malkowski, M.G. | | Deposit date: | 2022-12-06 | | Release date: | 2023-10-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Functional and structural analyses reveal that a dual domain sialidase protects bacteria from complement killing through desialylation of complement factors.
Plos Pathog., 19, 2023
|
|
6A73
 
 | | Complex structure of CSN2 with IP6 | | Descriptor: | COP9 signalosome complex subunit 2,Endolysin, INOSITOL HEXAKISPHOSPHATE, SULFATE ION | | Authors: | Liu, L, Li, D, Rao, F, Wang, T. | | Deposit date: | 2018-07-02 | | Release date: | 2019-07-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.447 Å) | | Cite: | Basis for metabolite-dependent Cullin-RING ligase deneddylation by the COP9 signalosome.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
7CE1
 
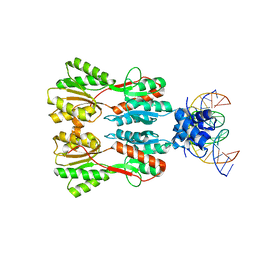 | | Complex STRUCTURE OF TRANSCRIPTION FACTOR SghR with its COGNATE DNA | | Descriptor: | LacI-type transcription factor, promoter DNA | | Authors: | Ye, F.Z, Wang, C, Yan, X.F, Zhang, L.H, Gao, Y.G. | | Deposit date: | 2020-06-21 | | Release date: | 2020-07-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural basis of a novel repressor, SghR, controllingAgrobacteriuminfection by cross-talking to plants.
J.Biol.Chem., 295, 2020
|
|
7CDV
 
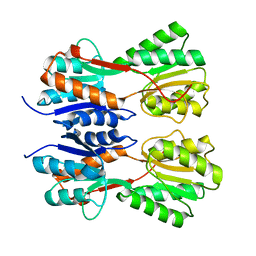 | | STRUCTURE OF A NOVEL VIRULENCE REGULATION FACTOR SghR | | Descriptor: | LacI-type transcription factor | | Authors: | Ye, F.Z, Wang, C, Yan, X.F, Zhang, L.H, Gao, Y.G. | | Deposit date: | 2020-06-20 | | Release date: | 2020-07-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis of a novel repressor, SghR, controllingAgrobacteriuminfection by cross-talking to plants.
J.Biol.Chem., 295, 2020
|
|
7CDX
 
 | | Complex STRUCTURE OF A NOVEL VIRULENCE REGULATION FACTOR SghR with its effector sucrose | | Descriptor: | LacI-type transcription factor, beta-D-fructofuranose-(2-1)-alpha-D-glucopyranose | | Authors: | Ye, F.Z, Wang, C, Yan, X.F, Zhang, L.H, Gao, Y.G. | | Deposit date: | 2020-06-20 | | Release date: | 2020-07-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.103 Å) | | Cite: | Structural basis of a novel repressor, SghR, controllingAgrobacteriuminfection by cross-talking to plants.
J.Biol.Chem., 295, 2020
|
|
7DH7
 
 | | Crystal structure of apo XcZur | | Descriptor: | PHOSPHATE ION, Transcriptional regulator fur family, ZINC ION | | Authors: | Liu, F.M, Su, Z.H, Chen, P, Tian, X.L, Wu, L.J, Tang, D.J, Li, P.F, Deng, H.T, Tang, J.L, Ming, Z.H. | | Deposit date: | 2020-11-13 | | Release date: | 2021-06-09 | | Last modified: | 2021-07-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis for zinc-induced activation of a zinc uptake transcriptional regulator.
Nucleic Acids Res., 49, 2021
|
|
7DH8
 
 | | Crystal structure of holo XcZur | | Descriptor: | Transcriptional regulator fur family, ZINC ION | | Authors: | Liu, F.M, Su, Z.H, Chen, P, Tian, X.L, Wu, L.J, Tang, D.J, Li, P.F, Deng, H.T, Tang, J.L, Ming, Z.H. | | Deposit date: | 2020-11-13 | | Release date: | 2021-06-09 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural basis for zinc-induced activation of a zinc uptake transcriptional regulator.
Nucleic Acids Res., 49, 2021
|
|
7YV4
 
 | | Crystal structure of human UCHL3 in complex with Farrerol | | Descriptor: | (2~{S})-2-(4-hydroxyphenyl)-6,8-dimethyl-5,7-bis(oxidanyl)-2,3-dihydrochromen-4-one, Ubiquitin carboxyl-terminal hydrolase isozyme L3 | | Authors: | Mao, Z.Y, Xu, X.J, Zhang, W.T. | | Deposit date: | 2022-08-18 | | Release date: | 2023-04-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Farrerol directly activates the deubiqutinase UCHL3 to promote DNA repair and reprogramming when mediated by somatic cell nuclear transfer.
Nat Commun, 14, 2023
|
|
