4OGT
 
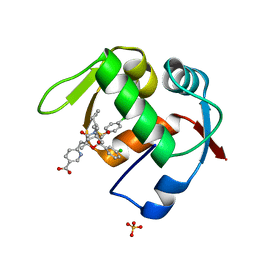 | | Co-Crystal Structure of MDM2 with Inhbitor Compound 46 | | Descriptor: | 6-{[(2R,5R,6R)-4-[(1S)-2-(tert-butylsulfonyl)-1-cyclopropylethyl]-6-(3-chlorophenyl)-5-(4-chlorophenyl)-2-methyl-3-oxomorpholin-2-yl]methyl}pyridine-3-carboxylic acid, E3 ubiquitin-protein ligase Mdm2, SULFATE ION | | Authors: | Shaffer, P.L, Huang, X, Yakowec, P, Long, A.M. | | Deposit date: | 2014-01-16 | | Release date: | 2014-04-02 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.5361 Å) | | Cite: | Novel Inhibitors of the MDM2-p53 Interaction Featuring Hydrogen Bond Acceptors as Carboxylic Acid Isosteres.
J.Med.Chem., 57, 2014
|
|
4OBA
 
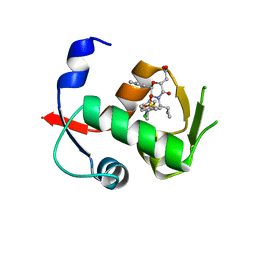 | | Co-crystal structure of MDM2 with Inhibitor Compound 4 | | Descriptor: | E3 ubiquitin-protein ligase Mdm2, [(2R,5R,6R)-4-[(1S)-2-(tert-butylsulfonyl)-1-cyclopropylethyl]-6-(3-chlorophenyl)-5-(4-chlorophenyl)-3-oxomorpholin-2-yl]acetic acid | | Authors: | Shaffer, P.L, Huang, X, Yakowec, P, Long, A.M. | | Deposit date: | 2014-01-07 | | Release date: | 2014-03-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.602 Å) | | Cite: | Selective and Potent Morpholinone Inhibitors of the MDM2-p53 Protein-Protein Interaction.
J.Med.Chem., 57, 2014
|
|
4OCC
 
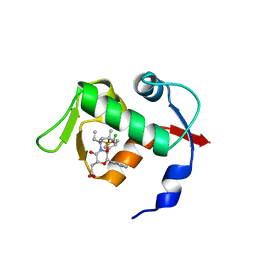 | | co-crystal structure of MDM2(17-111) in complex with compound 48 | | Descriptor: | E3 ubiquitin-protein ligase Mdm2, [(2R,5R,6R)-4-[(2S)-1-(tert-butylsulfonyl)butan-2-yl]-6-(3-chlorophenyl)-5-(4-chlorophenyl)-3-oxomorpholin-2-yl]acetic acid | | Authors: | Huang, X. | | Deposit date: | 2014-01-08 | | Release date: | 2014-04-02 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Novel Inhibitors of the MDM2-p53 Interaction Featuring Hydrogen Bond Acceptors as Carboxylic Acid Isosteres.
J.Med.Chem., 57, 2014
|
|
4ODF
 
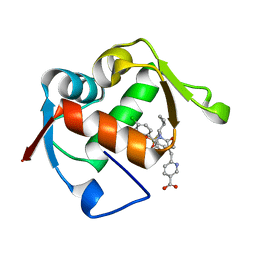 | | Co-Crystal Structure of MDM2 with Inhibitor Compound 47 | | Descriptor: | 6-{[(2S,5R,6R)-4-[(1S)-2-(tert-butylsulfonyl)-1-cyclopropylethyl]-6-(3-chlorophenyl)-5-(4-chlorophenyl)-2-methyl-3-oxomorpholin-2-yl]methyl}pyridine-3-carboxylic acid, E3 ubiquitin-protein ligase Mdm2 | | Authors: | Shaffer, P.L, Huang, X, Yakowec, P, Long, A.M. | | Deposit date: | 2014-01-10 | | Release date: | 2014-04-02 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.2006 Å) | | Cite: | Novel Inhibitors of the MDM2-p53 Interaction Featuring Hydrogen Bond Acceptors as Carboxylic Acid Isosteres.
J.Med.Chem., 57, 2014
|
|
6OAM
 
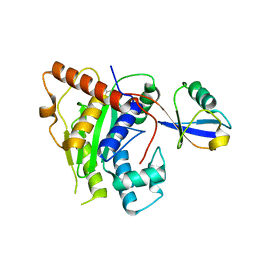 | | Crystal Structure of ChlaDUB2 DUB domain | | Descriptor: | Deubiquitinase and deneddylase Dub2, Ubiquitin | | Authors: | Hausman, J.M, Das, C. | | Deposit date: | 2019-03-17 | | Release date: | 2020-04-22 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.503 Å) | | Cite: | The Two Deubiquitinating Enzymes fromChlamydia trachomatisHave Distinct Ubiquitin Recognition Properties.
Biochemistry, 59, 2020
|
|
7VEY
 
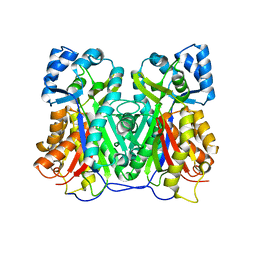 | | Crystal structure of Cyclosorus parasiticus chalcone synthase 1 (CpCHS1) | | Descriptor: | chalcone synthases | | Authors: | Li, J.X, Cheng, A.X. | | Deposit date: | 2021-09-10 | | Release date: | 2021-11-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Functional and Structural Investigation of Chalcone Synthases Based on Integrated Metabolomics and Transcriptome Analysis on Flavonoids and Anthocyanins Biosynthesis of the Fern Cyclosorus parasiticus .
Front Plant Sci, 12, 2021
|
|
7VEZ
 
 | |
7VF0
 
 | |
1I3Q
 
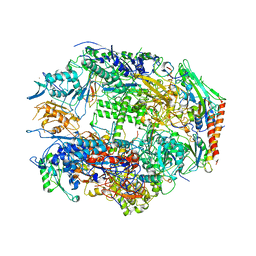 | | RNA POLYMERASE II CRYSTAL FORM I AT 3.1 A RESOLUTION | | Descriptor: | DNA-DIRECTED RNA POLYMERASE II 13.6KD POLYPEPTIDE, DNA-DIRECTED RNA POLYMERASE II 14.2KD POLYPEPTIDE, DNA-DIRECTED RNA POLYMERASE II 14.5KD POLYPEPTIDE, ... | | Authors: | Cramer, P, Bushnell, D.A, Kornberg, R.D. | | Deposit date: | 2001-02-15 | | Release date: | 2001-04-25 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural basis of transcription: RNA polymerase II at 2.8 angstrom resolution.
Science, 292, 2001
|
|
1I50
 
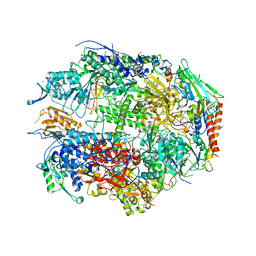 | | RNA POLYMERASE II CRYSTAL FORM II AT 2.8 A RESOLUTION | | Descriptor: | DNA-DIRECTED RNA POLYMERASE II 13.6KD POLYPEPTIDE, DNA-DIRECTED RNA POLYMERASE II 14.2KD POLYPEPTIDE, DNA-DIRECTED RNA POLYMERASE II 14.5KD POLYPEPTIDE, ... | | Authors: | Cramer, P, Bushnell, D.A, Kornberg, R.D. | | Deposit date: | 2001-02-23 | | Release date: | 2001-04-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis of transcription: RNA polymerase II at 2.8 angstrom resolution.
Science, 292, 2001
|
|
7CR2
 
 | | human KCNQ2 in complex with retigabine | | Descriptor: | Potassium voltage-gated channel subfamily KQT member 2, ethyl N-[2-azanyl-4-[(4-fluorophenyl)methylamino]phenyl]carbamate | | Authors: | Li, X, Lv, D, Wang, J, Ye, S, Guo, J. | | Deposit date: | 2020-08-12 | | Release date: | 2020-09-16 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Molecular basis for ligand activation of the human KCNQ2 channel.
Cell Res., 31, 2021
|
|
7CR7
 
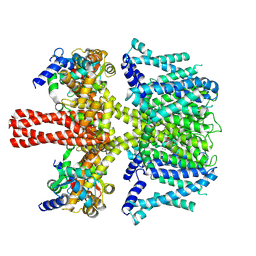 | | human KCNQ2-CaM in complex with retigabine | | Descriptor: | Calmodulin-3, Potassium voltage-gated channel subfamily KQT member 2, ethyl N-[2-azanyl-4-[(4-fluorophenyl)methylamino]phenyl]carbamate | | Authors: | Li, X, Lv, D, Wang, J, Ye, S, Guo, J. | | Deposit date: | 2020-08-12 | | Release date: | 2020-09-16 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Molecular basis for ligand activation of the human KCNQ2 channel.
Cell Res., 31, 2021
|
|
7CR0
 
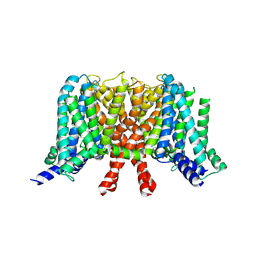 | | human KCNQ2 in apo state | | Descriptor: | Potassium voltage-gated channel subfamily KQT member 2 | | Authors: | Li, X, Lv, D, Wang, J, Ye, S, Guo, J. | | Deposit date: | 2020-08-12 | | Release date: | 2020-09-16 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Molecular basis for ligand activation of the human KCNQ2 channel.
Cell Res., 31, 2021
|
|
7CR3
 
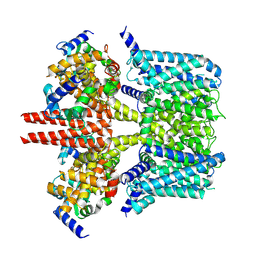 | | human KCNQ2-CaM in apo state | | Descriptor: | Calmodulin-3, Potassium voltage-gated channel subfamily KQT member 2 | | Authors: | Li, X, Lv, D, Wang, J, Ye, S, Guo, J. | | Deposit date: | 2020-08-12 | | Release date: | 2020-09-16 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Molecular basis for ligand activation of the human KCNQ2 channel.
Cell Res., 31, 2021
|
|
7CR1
 
 | | human KCNQ2 in complex with ztz240 | | Descriptor: | N-(6-chloranylpyridin-3-yl)-4-fluoranyl-benzamide, Potassium voltage-gated channel subfamily KQT member 2 | | Authors: | Li, X, Lv, D, Wang, J, Ye, S, Guo, J. | | Deposit date: | 2020-08-12 | | Release date: | 2020-09-16 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Molecular basis for ligand activation of the human KCNQ2 channel.
Cell Res., 31, 2021
|
|
7CR4
 
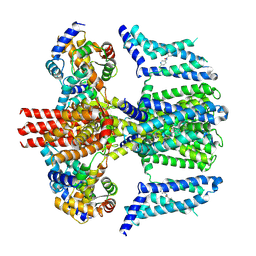 | | human KCNQ2-CaM in complex with ztz240 | | Descriptor: | Calmodulin-3, N-(6-chloranylpyridin-3-yl)-4-fluoranyl-benzamide, Potassium voltage-gated channel subfamily KQT member 2 | | Authors: | Li, X, Lv, D, Wang, J, Ye, S, Guo, J. | | Deposit date: | 2020-08-12 | | Release date: | 2020-09-16 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Molecular basis for ligand activation of the human KCNQ2 channel.
Cell Res., 31, 2021
|
|
6ULH
 
 | |
6UMS
 
 | |
6UMP
 
 | | Crystal structure of MavC in complex with substrate mimic in P65 space group | | Descriptor: | MavC, Ubiquitin, Ubiquitin-conjugating enzyme E2 N | | Authors: | Puvar, K, Iyer, S, Luo, Z.Q, Das, C. | | Deposit date: | 2019-10-10 | | Release date: | 2020-05-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Legionella effector MavC targets the Ube2N~Ub conjugate for noncanonical ubiquitination.
Nat Commun, 11, 2020
|
|
6BWE
 
 | | Sortase A from Corynebacterium diphtheriae, lid mutant | | Descriptor: | Putative fimbrial associated sortase-like protein | | Authors: | Osipiuk, J, Chang, C, Huang, I.H, Ton-That, H, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-12-14 | | Release date: | 2017-12-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | In vitro reconstitution of sortase-catalyzed pilus polymerization reveals structural elements involved in pilin cross-linking.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6P5H
 
 | |
6P5B
 
 | | Crystal Structure of MavC in Complex with Ub-UbE2N | | Descriptor: | MavC, Ubiquitin, Ubiquitin-conjugating enzyme E2 N | | Authors: | Puvar, K, Iyer, S, Negron Teron, K.I, Das, C. | | Deposit date: | 2019-05-30 | | Release date: | 2020-05-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.099 Å) | | Cite: | Legionella effector MavC targets the Ube2N~Ub conjugate for noncanonical ubiquitination.
Nat Commun, 11, 2020
|
|
6MAE
 
 | |
5Y70
 
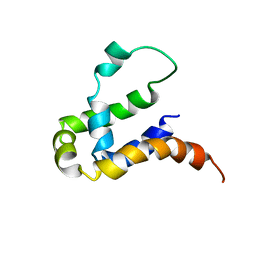 | | NMR structure of KMP11 in DPC micelle | | Descriptor: | Kinetoplastid membrane protein 11 | | Authors: | Lu, Y, Lim, L.Z, Song, J. | | Deposit date: | 2017-08-16 | | Release date: | 2018-02-07 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Kinetoplastid membrane protein-11 adopts a four-helix bundle fold in DPC micelle.
FEBS Lett., 591, 2017
|
|
5IE8
 
 | |
