7TPP
 
 | |
7TPQ
 
 | |
7UX1
 
 | | EcMscK in an Open Conformation | | Descriptor: | Mechanosensitive channel MscK | | Authors: | Mount, J.W, Yuan, P. | | Deposit date: | 2022-05-04 | | Release date: | 2022-11-23 | | Method: | ELECTRON MICROSCOPY (3.48 Å) | | Cite: | Structural basis for mechanotransduction in a potassium-dependent mechanosensitive ion channel.
Nat Commun, 13, 2022
|
|
7UW5
 
 | | EcMscK G924S mutant in a closed conformation | | Descriptor: | Mechanosensitive channel MscK | | Authors: | Mount, J.W, Yuan, P. | | Deposit date: | 2022-05-02 | | Release date: | 2022-11-23 | | Method: | ELECTRON MICROSCOPY (3.84 Å) | | Cite: | Structural basis for mechanotransduction in a potassium-dependent mechanosensitive ion channel.
Nat Commun, 13, 2022
|
|
8F7C
 
 | | Cryo-EM structure of human pannexin 2 | | Descriptor: | Pannexin-2, Soluble cytochrome b562 fusion | | Authors: | He, Z, Yuan, P. | | Deposit date: | 2022-11-18 | | Release date: | 2023-04-05 | | Method: | ELECTRON MICROSCOPY (3.92 Å) | | Cite: | Structural and functional analysis of human pannexin 2 channel.
Nat Commun, 14, 2023
|
|
8F35
 
 | | Apo ELIC in spMSP1D1 nanodiscs with 2:1:1 POPC:POPE:POPG | | Descriptor: | Erwinia chrysanthemi ligand-gated ion channel | | Authors: | Dalal, V, Arcario, M.J, Petroff II, J.T, Deitzen, N.M, Tan, B.K, Brannigan, G, Cheng, W.W.L. | | Deposit date: | 2022-11-09 | | Release date: | 2023-11-15 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.17 Å) | | Cite: | Lipid nanodisc scaffold and size alter the structure of a pentameric ligand-gated ion channel.
Nat Commun, 15, 2024
|
|
8F32
 
 | | ELIC with Propylamine in SMA nanodiscs with 2:1:1 POPC:POPE:POPG | | Descriptor: | 3-AMINOPROPANE, Erwinia chrysanthemi ligand-gated ion channel | | Authors: | Dalal, V, Arcario, M.J, Petroff II, J.T, Deitzen, N.M, Tan, B.K, Brannigan, G, Cheng, W.W.L. | | Deposit date: | 2022-11-09 | | Release date: | 2023-11-15 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.71 Å) | | Cite: | Lipid nanodisc scaffold and size alter the structure of a pentameric ligand-gated ion channel.
Nat Commun, 15, 2024
|
|
8F34
 
 | | ELIC with Propylamine in spMSP1D1 nanodiscs with 2:1:1 POPC:POPE:POPG | | Descriptor: | 3-AMINOPROPANE, Erwinia chrysanthemi ligand-gated ion channel | | Authors: | Dalal, V, Arcario, M.J, Petroff II, J.T, Deitzen, N.M, Tan, B.K, Brannigan, G, Cheng, W.W.L. | | Deposit date: | 2022-11-09 | | Release date: | 2023-11-15 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.12 Å) | | Cite: | Lipid nanodisc scaffold and size alter the structure of a pentameric ligand-gated ion channel.
Nat Commun, 15, 2024
|
|
8F33
 
 | | ELIC with Propylamine in saposin nanodiscs with 2:1:1 POPC:POPE:POPG | | Descriptor: | 3-AMINOPROPANE, Erwinia chrysanthemi ligand-gated ion channel | | Authors: | Dalal, V, Arcario, M.J, Petroff II, J.T, Deitzen, N.M, Tan, B.K, Brannigan, G, Cheng, W.W.L. | | Deposit date: | 2022-11-09 | | Release date: | 2023-11-15 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.28 Å) | | Cite: | Lipid nanodisc scaffold and size alter the structure of a pentameric ligand-gated ion channel.
Nat Commun, 15, 2024
|
|
7K9J
 
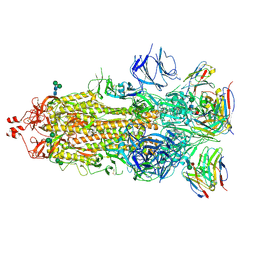 | |
7K9I
 
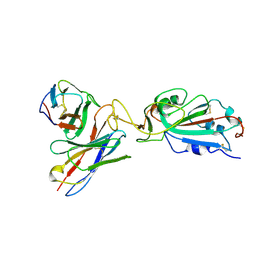 | |
7K9K
 
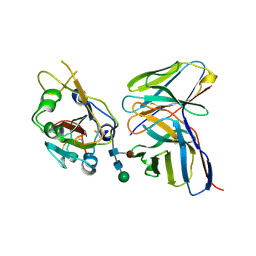 | |
7K9H
 
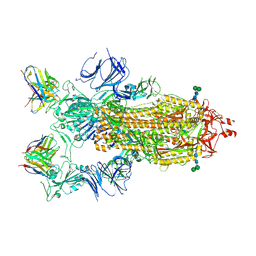 | | SARS-CoV-2 Spike in complex with neutralizing Fab 2B04 (one up, two down conformation) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2B04 heavy chain, ... | | Authors: | Errico, J.M, Fremont, D.H, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-09-29 | | Release date: | 2021-09-29 | | Last modified: | 2021-11-10 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural mechanism of SARS-CoV-2 neutralization by two murine antibodies targeting the RBD.
Cell Rep, 37, 2021
|
|
7KVE
 
 | |
7KXY
 
 | |
7KUI
 
 | | Cryo-EM structure of Rous sarcoma virus cleaved synaptic complex (CSC) with HIV-1 integrase strand transfer inhibitor MK-2048. CIC region of a cluster identified by 3-dimensional variability analysis in cryoSPARC. | | Descriptor: | (6S)-2-(3-chloro-4-fluorobenzyl)-8-ethyl-10-hydroxy-N,6-dimethyl-1,9-dioxo-1,2,6,7,8,9-hexahydropyrazino[1',2':1,5]pyrrolo[2,3-d]pyridazine-4-carboxamide, DNA (5'-D(*AP*AP*TP*GP*TP*TP*GP*TP*CP*TP*TP*AP*TP*GP*CP*AP*AP*T)-3'), DNA (5'-D(*AP*TP*TP*GP*CP*AP*TP*AP*AP*GP*AP*CP*AP*AP*CP*A)-3'), ... | | Authors: | Pandey, K.K, Bera, S, Shi, K, Aihara, H, Grandgenett, D.P. | | Deposit date: | 2020-11-25 | | Release date: | 2021-03-17 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-EM structure of the Rous sarcoma virus octameric cleaved synaptic complex intasome.
Commun Biol, 4, 2021
|
|
7KU7
 
 | | Cryo-EM structure of Rous sarcoma virus cleaved synaptic complex (CSC) with HIV-1 integrase strand transfer inhibitor MK-2048. Cluster identified by 3-dimensional variability analysis in cryoSPARC. | | Descriptor: | (6S)-2-(3-chloro-4-fluorobenzyl)-8-ethyl-10-hydroxy-N,6-dimethyl-1,9-dioxo-1,2,6,7,8,9-hexahydropyrazino[1',2':1,5]pyrrolo[2,3-d]pyridazine-4-carboxamide, DNA (5'-D(*AP*AP*TP*GP*TP*TP*GP*TP*CP*TP*TP*AP*TP*GP*CP*AP*AP*T)-3'), DNA (5'-D(*AP*TP*TP*GP*CP*AP*TP*AP*AP*GP*AP*CP*AP*AP*CP*A)-3'), ... | | Authors: | Pandey, K.K, Bera, S, Shi, K, Aihara, H, Grandgenett, D.P. | | Deposit date: | 2020-11-24 | | Release date: | 2021-03-17 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-EM structure of the Rous sarcoma virus octameric cleaved synaptic complex intasome.
Commun Biol, 4, 2021
|
|
7KVF
 
 | |
