2H0Z
 
 | |
2HO7
 
 | |
2HOK
 
 | |
2HO6
 
 | |
2HOM
 
 | |
2HOL
 
 | |
2HOJ
 
 | |
2HOP
 
 | |
2HOO
 
 | |
6E1V
 
 | | Crystal structure of a class I PreQ1 riboswitch complexed with a synthetic compound 3: 2-[(9H-carbazol-3-yl)oxy]-N,N-dimethylethan-1-amine | | Descriptor: | 2-[(9H-carbazol-3-yl)oxy]-N,N-dimethylethan-1-amine, RNA (33-MER) | | Authors: | Numata, T, Connelly, C.M, Schneekloth, J.S, Ferre-D'Amare, A.R. | | Deposit date: | 2018-07-10 | | Release date: | 2019-04-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Synthetic ligands for PreQ1riboswitches provide structural and mechanistic insights into targeting RNA tertiary structure.
Nat Commun, 10, 2019
|
|
6E1S
 
 | | Crystal structure of a class I PreQ1 riboswitch complexed with a synthetic compound 1: 2-[(dibenzo[b,d]furan-2-yl)oxy]ethan-1-amine | | Descriptor: | 2-[(dibenzo[b,d]furan-2-yl)oxy]ethan-1-amine, RNA (33-MER) | | Authors: | Numata, T, Connelly, C.M, Schneekloth, J.S, Ferre-D'Amare, A.R. | | Deposit date: | 2018-07-10 | | Release date: | 2019-04-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Synthetic ligands for PreQ1riboswitches provide structural and mechanistic insights into targeting RNA tertiary structure.
Nat Commun, 10, 2019
|
|
6E1T
 
 | | Crystal structure of a class I PreQ1 riboswitch complexed with a synthetic compound 1: 2-[(dibenzo[b,d]furan-2-yl)oxy]ethan-1-amine | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-[(dibenzo[b,d]furan-2-yl)oxy]ethan-1-amine, MAGNESIUM ION, ... | | Authors: | Numata, T, Connelly, C.M, Schneekloth, J.S, Ferre-D'Amare, A.R. | | Deposit date: | 2018-07-10 | | Release date: | 2019-04-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Synthetic ligands for PreQ1riboswitches provide structural and mechanistic insights into targeting RNA tertiary structure.
Nat Commun, 10, 2019
|
|
6E1W
 
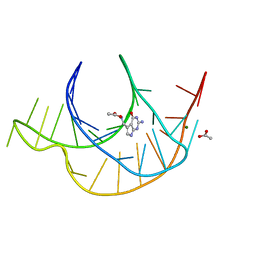 | | Crystal structure of a class I PreQ1 riboswitch complexed with PreQ1 | | Descriptor: | 2-amino-5-(aminomethyl)-1,7-dihydro-4H-pyrrolo[2,3-d]pyrimidin-4-one, ACETATE ION, MAGNESIUM ION, ... | | Authors: | Numata, T, Connelly, C.M, Schneekloth, J.S, Ferre-D'Amare, A.R. | | Deposit date: | 2018-07-10 | | Release date: | 2019-04-10 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Synthetic ligands for PreQ1riboswitches provide structural and mechanistic insights into targeting RNA tertiary structure.
Nat Commun, 10, 2019
|
|
6E1U
 
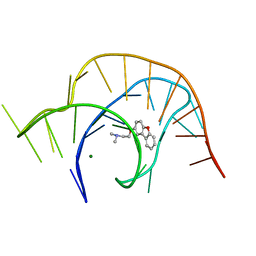 | | Crystal structure of a class I PreQ1 riboswitch complexed with a synthetic compound 2: 2-[(dibenzo[b,d]furan-2-yl)oxy]-N,N-dimethylethan-1-amine | | Descriptor: | 2-[(dibenzo[b,d]furan-2-yl)oxy]-N,N-dimethylethan-1-amine, MAGNESIUM ION, RNA (33-MER) | | Authors: | Numata, T, Connelly, C.M, Schneekloth, J.S, Ferre-D'Amare, A.R. | | Deposit date: | 2018-07-10 | | Release date: | 2019-04-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Synthetic ligands for PreQ1riboswitches provide structural and mechanistic insights into targeting RNA tertiary structure.
Nat Commun, 10, 2019
|
|
6E8T
 
 | |
6E8S
 
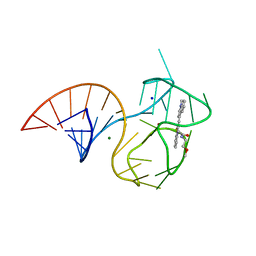 | | Structure of the iMango-III aptamer bound to TO1-Biotin | | Descriptor: | 4-[(3-{2-[(2-methoxyethyl)amino]-2-oxoethyl}-1,3-benzothiazol-3-ium-2-yl)methyl]-1-methylquinolin-1-ium, MAGNESIUM ION, POTASSIUM ION, ... | | Authors: | Trachman, R.J, Ferre-D'Amare, A.R. | | Deposit date: | 2018-07-31 | | Release date: | 2019-04-17 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structure and functional reselection of the Mango-III fluorogenic RNA aptamer.
Nat. Chem. Biol., 15, 2019
|
|
6E8U
 
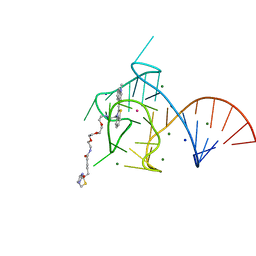 | | Structure of the Mango-III (A10U) aptamer bound to TO1-Biotin | | Descriptor: | 4-[(3-{2,16-dioxo-20-[(3aR,4R,6aS)-2-oxohexahydro-1H-thieno[3,4-d]imidazol-4-yl]-6,9,12-trioxa-3,15-diazaicosan-1-yl}-1,3-benzothiazol-3-ium-2-yl)methyl]-1-methylquinolin-1-ium, MAGNESIUM ION, POTASSIUM ION, ... | | Authors: | Trachman, R.J, Ferre-D'Amare, A.R. | | Deposit date: | 2018-07-31 | | Release date: | 2019-04-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structure and functional reselection of the Mango-III fluorogenic RNA aptamer.
Nat. Chem. Biol., 15, 2019
|
|
6E80
 
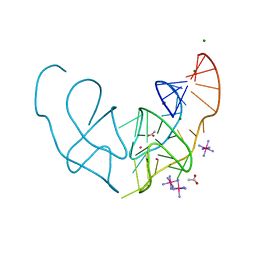 | |
6E84
 
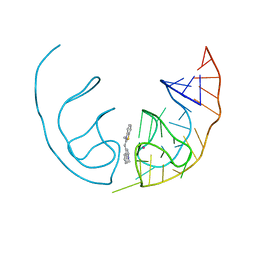 | | Crystal structure of the Corn aptamer in complex with TO | | Descriptor: | 1-methyl-4-[(Z)-(3-methyl-1,3-benzothiazol-2(3H)-ylidene)methyl]quinolin-1-ium, POTASSIUM ION, RNA (36-MER) | | Authors: | Sjekloca, L, Ferre-D'Amare, A.R. | | Deposit date: | 2018-07-27 | | Release date: | 2019-07-31 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.901 Å) | | Cite: | Binding between G Quadruplexes at the Homodimer Interface of the Corn RNA Aptamer Strongly Activates Thioflavin T Fluorescence.
Cell Chem Biol, 26, 2019
|
|
6E82
 
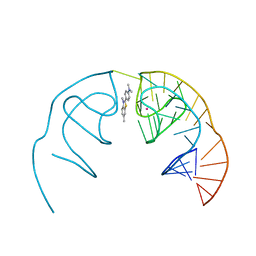 | |
6E81
 
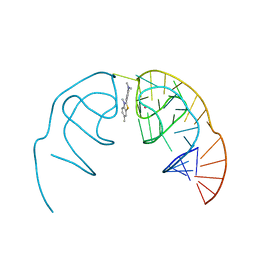 | | Crystal structure of the Corn aptamer in complex with ThT | | Descriptor: | 2-[4-(dimethylamino)phenyl]-3,6-dimethyl-1,3-benzothiazol-3-ium, POTASSIUM ION, RNA (36-MER) | | Authors: | Sjekloca, L, Ferre-D'Amare, A.R. | | Deposit date: | 2018-07-27 | | Release date: | 2019-07-31 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.721 Å) | | Cite: | Binding between G Quadruplexes at the Homodimer Interface of the Corn RNA Aptamer Strongly Activates Thioflavin T Fluorescence.
Cell Chem Biol, 26, 2019
|
|
3OT0
 
 | |
6PUP
 
 | |
6OD9
 
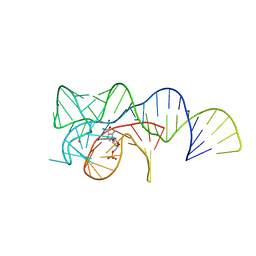 | | Co-crystal structure of the Fusobacterium ulcerans ZTP riboswitch using an X-ray free-electron laser | | Descriptor: | AMINOIMIDAZOLE 4-CARBOXAMIDE RIBONUCLEOTIDE, CESIUM ION, MAGNESIUM ION, ... | | Authors: | Jones, C.P, Tran, B, Ferre-D'Amare, A.R. | | Deposit date: | 2019-03-26 | | Release date: | 2019-07-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (4.102 Å) | | Cite: | Co-crystal structure of the Fusobacterium ulcerans ZTP riboswitch using an X-ray free-electron laser.
Acta Crystallogr.,Sect.F, 75, 2019
|
|
1AM9
 
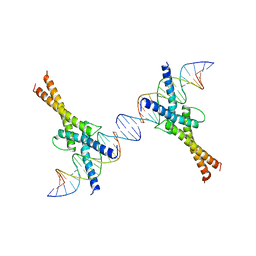 | | HUMAN SREBP-1A BOUND TO LDL RECEPTOR PROMOTER | | Descriptor: | DNA (5'-D(*CP*AP*TP*GP*AP*GP*AP*TP*CP*AP*CP*CP*CP*CP*AP*CP*T P*GP*CP*AP*A)-3'), DNA (5'-D(*TP*TP*GP*CP*AP*GP*TP*GP*GP*GP*GP*TP*GP*AP*TP*CP*T )-3'), MAGNESIUM ION, ... | | Authors: | Parraga, A, Burley, S.K. | | Deposit date: | 1997-06-25 | | Release date: | 1998-07-10 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Co-crystal structure of sterol regulatory element binding protein 1a at 2.3 A resolution.
Structure, 6, 1998
|
|
