4NZW
 
 | | Crystal Structure of STK25-MO25 Complex | | 分子名称: | 5-[(E)-(5-CHLORO-2-OXO-1,2-DIHYDRO-3H-INDOL-3-YLIDENE)METHYL]-N-[2-(DIETHYLAMINO)ETHYL]-2,4-DIMETHYL-1H-PYRROLE-3-CARBOXAMIDE, Calcium-binding protein 39, Serine/threonine-protein kinase 25 | | 著者 | Feng, M, Hao, Q, Zhou, Z.C. | | 登録日 | 2013-12-13 | | 公開日 | 2014-12-03 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (3.583 Å) | | 主引用文献 | Structural insights into regulatory mechanisms of MO25-mediated kinase activation.
J.Struct.Biol., 186, 2014
|
|
7LKJ
 
 | | Crystal structure of Helicobacter pylori aminofutalosine deaminase (AFLDA) | | 分子名称: | 1,2-ETHANEDIOL, Aminofutalosine deaminase, FE (III) ION | | 著者 | Harijan, R.K, Feng, M, Bonanno, J.B, Almo, S.C, Schramm, V.L. | | 登録日 | 2021-02-02 | | 公開日 | 2021-06-23 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.79 Å) | | 主引用文献 | Aminofutalosine Deaminase in the Menaquinone Pathway of Helicobacter pylori .
Biochemistry, 60, 2021
|
|
7LKK
 
 | | Crystal structure of Helicobacter pylori aminofutalosine deaminase (AFLDA) in complex with Methylthio-coformycin | | 分子名称: | (8R)-3-(5-S-methyl-5-thio-beta-D-ribofuranosyl)-3,6,7,8-tetrahydroimidazo[4,5-d][1,3]diazepin-8-ol, 1,2-ETHANEDIOL, Aminofutalosine deaminase, ... | | 著者 | Harijan, R.K, Feng, M, Bonanno, J.B, Almo, S.C, Schramm, V.L. | | 登録日 | 2021-02-02 | | 公開日 | 2021-06-23 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.89 Å) | | 主引用文献 | Aminofutalosine Deaminase in the Menaquinone Pathway of Helicobacter pylori .
Biochemistry, 60, 2021
|
|
4O27
 
 | | Crystal structure of MST3-MO25 complex with WIF motif | | 分子名称: | 5-mer peptide from serine/threonine-protein kinase 24, ADENOSINE-5'-DIPHOSPHATE, Calcium-binding protein 39, ... | | 著者 | Hao, Q, Feng, M, Zhou, Z.C. | | 登録日 | 2013-12-16 | | 公開日 | 2014-12-03 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (3.185 Å) | | 主引用文献 | Structural insights into regulatory mechanisms of MO25-mediated kinase activation.
J.Struct.Biol., 186, 2014
|
|
5HNK
 
 | | Crystal structure of T5Fen in complex intact substrate and metal ions. | | 分子名称: | DNA (5'-D(*AP*AP*AP*AP*GP*CP*GP*TP*AP*CP*GP*C)-3'), Exodeoxyribonuclease, GLYCEROL, ... | | 著者 | Almalki, F.A, Feng, M, Zhang, J, Sedelnikova, S.E, Rafferty, J.B, Sayers, J.R, Artymiuk, P.J. | | 登録日 | 2016-01-18 | | 公開日 | 2016-06-01 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.22 Å) | | 主引用文献 | Direct observation of DNA threading in flap endonuclease complexes.
Nat.Struct.Mol.Biol., 23, 2016
|
|
5HML
 
 | | Crystal Structure of T5 D15 Protein Co-crystallized with Metal Ions | | 分子名称: | 1,2-ETHANEDIOL, 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, ... | | 著者 | Flemming, C.S, Feng, M, Sedelnikova, S.E, Zhang, J, Rafferty, J.B, Sayers, J.R, Artymiuk, P.J. | | 登録日 | 2016-01-16 | | 公開日 | 2016-06-01 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.482 Å) | | 主引用文献 | Direct observation of DNA threading in flap endonuclease complexes.
Nat.Struct.Mol.Biol., 23, 2016
|
|
1UT5
 
 | |
1UT8
 
 | |
3SJL
 
 | |
3SLE
 
 | |
3SVW
 
 | |
6XIG
 
 | |
6XI9
 
 | | X-ray crystal structure of MqnE from Pedobacter heparinus in complex with aminofutalosine and methionine | | 分子名称: | 9-[7-(3-carboxyphenyl)-5,6-dideoxy-beta-D-ribo-heptodialdo-1,4-furanosyl]-9H-purin-6-amine, Aminodeoxyfutalosine synthase, CHLORIDE ION, ... | | 著者 | Grove, T.L, Bonanno, J.B, Almo, S.C. | | 登録日 | 2020-06-19 | | 公開日 | 2020-07-15 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (2.14 Å) | | 主引用文献 | Narrow-Spectrum Antibiotic Targeting of the Radical SAM Enzyme MqnE in Menaquinone Biosynthesis.
Biochemistry, 59, 2020
|
|
6S6B
 
 | | Type III-B Cmr-beta Cryo-EM structure of the Apo state | | 分子名称: | CRISPR-associated RAMP protein, Cmr4 family, Cmr6 family, ... | | 著者 | Sofos, N, Montoya, G, Stella, S. | | 登録日 | 2019-07-02 | | 公開日 | 2020-07-08 | | 最終更新日 | 2020-09-16 | | 実験手法 | ELECTRON MICROSCOPY (2.75 Å) | | 主引用文献 | Structures of the Cmr-beta Complex Reveal the Regulation of the Immunity Mechanism of Type III-B CRISPR-Cas.
Mol.Cell, 79, 2020
|
|
6S8B
 
 | | Cryo-EM structure of the Type III-B Cmr-beta bound to cognate target RNA and AMPPnP, state 1 | | 分子名称: | CRISPR-associated RAMP protein, Cmr4 family, Cmr6 family, ... | | 著者 | Sofos, N, Montoya, G, Stella, S. | | 登録日 | 2019-07-09 | | 公開日 | 2020-07-08 | | 最終更新日 | 2020-09-16 | | 実験手法 | ELECTRON MICROSCOPY (2.41 Å) | | 主引用文献 | Structures of the Cmr-beta Complex Reveal the Regulation of the Immunity Mechanism of Type III-B CRISPR-Cas.
Mol.Cell, 79, 2020
|
|
6S91
 
 | | Cryo-EM structure of the Type III-B Cmr-beta bound to cognate target RNA and AMPPnP, state 2 | | 分子名称: | CRISPR-associated RAMP protein, Cmr4 family, Cmr6 family, ... | | 著者 | Sofos, N, Montoya, G, Stella, S. | | 登録日 | 2019-07-11 | | 公開日 | 2020-07-08 | | 最終更新日 | 2020-09-16 | | 実験手法 | ELECTRON MICROSCOPY (2.68 Å) | | 主引用文献 | Structures of the Cmr-beta Complex Reveal the Regulation of the Immunity Mechanism of Type III-B CRISPR-Cas.
Mol.Cell, 79, 2020
|
|
6SIC
 
 | | Cryo-EM structure of the Type III-B Cmr-beta bound to cognate target RNA | | 分子名称: | CRISPR-associated RAMP protein, Cmr1 family, Cmr4 family, ... | | 著者 | Sofos, N, Montoya, G, Stella, S. | | 登録日 | 2019-08-09 | | 公開日 | 2020-07-08 | | 最終更新日 | 2020-09-16 | | 実験手法 | ELECTRON MICROSCOPY (3.52 Å) | | 主引用文献 | Structures of the Cmr-beta Complex Reveal the Regulation of the Immunity Mechanism of Type III-B CRISPR-Cas.
Mol.Cell, 79, 2020
|
|
6S8E
 
 | | Cryo-EM structure of the type III-B Cmr-beta complex bound to non-cognate target RNA | | 分子名称: | CRISPR-associated RAMP protein, Cmr4 family, Cmr6 family, ... | | 著者 | Sofos, N, Montoya, G, Stella, S. | | 登録日 | 2019-07-09 | | 公開日 | 2020-07-08 | | 最終更新日 | 2020-09-16 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | Structures of the Cmr-beta Complex Reveal the Regulation of the Immunity Mechanism of Type III-B CRISPR-Cas.
Mol.Cell, 79, 2020
|
|
6SH8
 
 | | Cryo-EM structure of the Type III-B Cmr-beta bound to cognate target RNA and AMPPnP, state 2, in the presence of ssDNA | | 分子名称: | CRISPR-associated RAMP protein, Cmr4 family, Cmr6 family, ... | | 著者 | Sofos, N, Montoya, G, Stella, S. | | 登録日 | 2019-08-06 | | 公開日 | 2020-07-08 | | 最終更新日 | 2020-09-16 | | 実験手法 | ELECTRON MICROSCOPY (3.14 Å) | | 主引用文献 | Structures of the Cmr-beta Complex Reveal the Regulation of the Immunity Mechanism of Type III-B CRISPR-Cas.
Mol.Cell, 79, 2020
|
|
6SHB
 
 | | Cryo-EM structure of the Type III-B Cmr-beta bound to cognate target RNA and AMPPnP, state 1, in the presence of ssDNA | | 分子名称: | CRISPR-associated RAMP protein, Cmr4 family, Cmr6 family, ... | | 著者 | Sofos, N, Montoya, G, Stella, S. | | 登録日 | 2019-08-06 | | 公開日 | 2020-07-08 | | 最終更新日 | 2020-09-16 | | 実験手法 | ELECTRON MICROSCOPY (3.07 Å) | | 主引用文献 | Structures of the Cmr-beta Complex Reveal the Regulation of the Immunity Mechanism of Type III-B CRISPR-Cas.
Mol.Cell, 79, 2020
|
|
4Y5R
 
 | | Crystal Structure of a T67A MauG/pre-Methylamine Dehydrogenase Complex | | 分子名称: | CALCIUM ION, HEME C, Methylamine dehydrogenase heavy chain, ... | | 著者 | Li, C, Wilmot, C.M. | | 登録日 | 2015-02-11 | | 公開日 | 2015-06-17 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | A T67A mutation in the proximal pocket of the high-spin heme of MauG stabilizes formation of a mixed-valent Fe(II)/Fe(III) state and enhances charge resonance stabilization of the bis-Fe(IV) state.
Biochim.Biophys.Acta, 1847, 2015
|
|
6QZQ
 
 | |
6QZT
 
 | |
6R9R
 
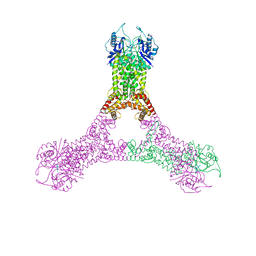 | | Crystal structure of Csx1 in complex with cyclic oligoadenylate cOA4 conformation 2 | | 分子名称: | CRISPR-associated (Cas) DxTHG family, circular RNA (5'-R(P*AP*AP*AP*A)-3') | | 著者 | Molina, R, Montoya, G, Sofos, N, Stella, S. | | 登録日 | 2019-04-03 | | 公開日 | 2019-10-02 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Structure of Csx1-cOA4complex reveals the basis of RNA decay in Type III-B CRISPR-Cas.
Nat Commun, 10, 2019
|
|
6R7B
 
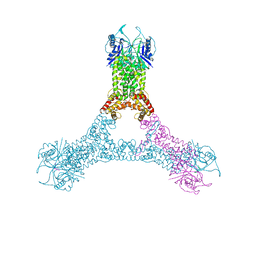 | | Crystal structure of Csx1 in complex with cyclic oligoadenylate cOA4 conformation 1 | | 分子名称: | CRISPR-associated (Cas) DxTHG family, RNA (5'-R(P*AP*AP*AP*A)-3') | | 著者 | Molina, R, Montoya, G, Sofos, N, Stella, S. | | 登録日 | 2019-03-28 | | 公開日 | 2019-10-02 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (3.12 Å) | | 主引用文献 | Structure of Csx1-cOA4complex reveals the basis of RNA decay in Type III-B CRISPR-Cas.
Nat Commun, 10, 2019
|
|
