7DVW
 
 | | SARS-CoV-2 Mpro mutant (H41A) in complex with nsp5|6 peptidyl substrate | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE, nsp5/6 peptidyl substrate | | Authors: | Liu, X, Zhao, Y, Yang, H, Rao, Z. | | Deposit date: | 2021-01-15 | | Release date: | 2022-01-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Structural basis for replicase polyprotein cleavage and substrate specificity of main protease from SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7DVY
 
 | | SARS-CoV-2 Mpro mutant (H41A) in complex with nsp9|10 peptidyl substrate | | Descriptor: | 3C-like proteinase, nsp9/10 peptidyl substrate | | Authors: | Liu, X, Zhao, Y, Yang, H, Rao, Z. | | Deposit date: | 2021-01-15 | | Release date: | 2022-01-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for replicase polyprotein cleavage and substrate specificity of main protease from SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7DVX
 
 | | SARS-CoV-2 Mpro mutant (H41A) in complex with nsp6|7 peptidyl substrate | | Descriptor: | 3C-like proteinase, nsp6/7 peptidyl substrate | | Authors: | Liu, X, Zhao, Y, Yang, H, Rao, Z. | | Deposit date: | 2021-01-15 | | Release date: | 2022-01-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for replicase polyprotein cleavage and substrate specificity of main protease from SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7E5X
 
 | |
4QNC
 
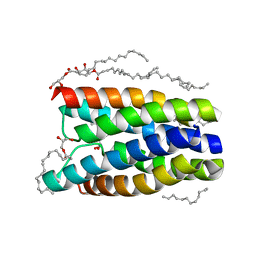 | | Crystal structure of a SemiSWEET in an occluded state | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, PENTADECANE, chemical transport protein | | Authors: | Yan, X, Yuyong, T, Liang, F, Perry, K. | | Deposit date: | 2014-06-17 | | Release date: | 2014-09-10 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.388 Å) | | Cite: | Structures of bacterial homologues of SWEET transporters in two distinct conformations.
Nature, 515, 2014
|
|
7W11
 
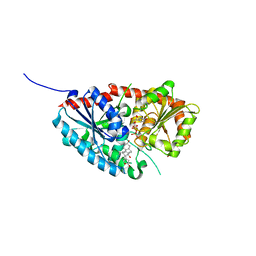 | | glycosyltransferase | | Descriptor: | DIGITOXIGENIN, Glycosyltransferase, URIDINE-5'-DIPHOSPHATE, ... | | Authors: | Wei, H, Feng, L. | | Deposit date: | 2021-11-19 | | Release date: | 2022-03-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Functional and Structural Dissection of a Plant Steroid 3-O-Glycosyltransferase Facilitated the Engineering Enhancement of Sugar Donor Promiscuity
Acs Catalysis, 2022
|
|
7W09
 
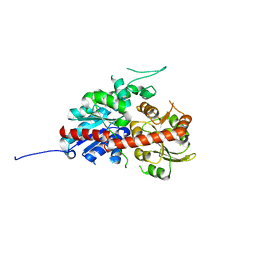 | | UGT74AN2, Plant Steroid Glycosyltransferase | | Descriptor: | Glycosyltransferase | | Authors: | Wei, H, Feng, L. | | Deposit date: | 2021-11-18 | | Release date: | 2022-03-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Functional and Structural Dissection of a Plant Steroid 3-O-Glycosyltransferase Facilitated the Engineering Enhancement of Sugar Donor Promiscuity
Acs Catalysis, 2022
|
|
7W1B
 
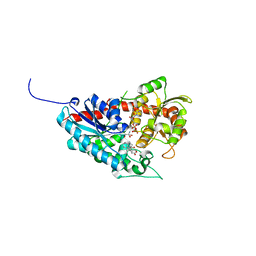 | | Glycosyltransferase | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DIGITOXIGENIN, Glycosyltransferase, ... | | Authors: | Wei, H, Feng, L. | | Deposit date: | 2021-11-19 | | Release date: | 2022-03-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Functional and Structural Dissection of a Plant Steroid 3-O-Glycosyltransferase Facilitated the Engineering Enhancement of Sugar Donor Promiscuity
Acs Catalysis, 2022
|
|
7W0K
 
 | | plant glycosyltransferase | | Descriptor: | GLYCEROL, Glycosyltransferase, URIDINE-5'-DIPHOSPHATE | | Authors: | Wei, H, Feng, L. | | Deposit date: | 2021-11-18 | | Release date: | 2022-03-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Functional and Structural Dissection of a Plant Steroid 3-O-Glycosyltransferase Facilitated the Engineering Enhancement of Sugar Donor Promiscuity
Acs Catalysis, 2022
|
|
7W1H
 
 | | glycosyltransferase | | Descriptor: | Glycosyltransferase, URIDINE-5'-DIPHOSPHATE-GLUCOSE | | Authors: | Wei, H, Feng, L. | | Deposit date: | 2021-11-19 | | Release date: | 2022-03-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Functional and Structural Dissection of a Plant Steroid 3-O-Glycosyltransferase Facilitated the Engineering Enhancement of Sugar Donor Promiscuity
Acs Catalysis, 2022
|
|
7W0Z
 
 | | Glycosyltranferase UGT74AN2 | | Descriptor: | 5-[(1R,2S,4R,6R,7R,10S,11S,14S,16R)-14-hydroxy-7,11-dimethyl-3-oxapentacyclo[8.8.0.02,4.02,7.011,16]octadecan-6-yl]pyran-2-one, Glycosyltransferase, URIDINE-5'-DIPHOSPHATE | | Authors: | Wei, H, Feng, L. | | Deposit date: | 2021-11-18 | | Release date: | 2022-03-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Functional and Structural Dissection of a Plant Steroid 3-O-Glycosyltransferase Facilitated the Engineering Enhancement of Sugar Donor Promiscuity
Acs Catalysis, 2022
|
|
5XXY
 
 | |
5Y8V
 
 | | Crystal structure of GAS41 | | Descriptor: | MAGNESIUM ION, YEATS domain-containing protein 4 | | Authors: | Wang, Y, Hao, Q. | | Deposit date: | 2017-08-21 | | Release date: | 2018-04-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Identification of the YEATS domain of GAS41 as a pH-dependent reader of histone succinylation
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
5YJ5
 
 | |
5YJ4
 
 | |
1QFD
 
 | | NMR SOLUTION STRUCTURE OF ALPHA-AMYLASE INHIBITOR (AAI) | | Descriptor: | PROTEIN (ALPHA-AMYLASE INHIBITOR) | | Authors: | Lu, S, Deng, P, Liu, X, Luo, J, Han, R, Gu, X, Liang, S, Wang, X, Feng, L, Lozanov, V, Patthy, A, Pongor, S. | | Deposit date: | 1999-04-08 | | Release date: | 1999-07-16 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the major alpha-amylase inhibitor of the crop plant amaranth.
J.Biol.Chem., 274, 1999
|
|
7XOG
 
 | | Cryo-EM structure of S glycoprotein encoded by the Covid-19 mRNA vaccine candidate RQ3013 (Postfusion state) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein,peptide, ... | | Authors: | Wu, Z, Yu, Z, Tan, S, Lu, J, Lu, G, Lin, J. | | Deposit date: | 2022-05-01 | | Release date: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Preclinical evaluation of RQ3013, a broad-spectrum mRNA vaccine against SARS-CoV-2 variants.
Sci Bull (Beijing), 68, 2023
|
|
7XOE
 
 | | Cryo-EM structure of S glycoprotein encoded by the Covid-19 mRNA vaccine candidate RQ3013 (Prefusion state) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein,peptide | | Authors: | Wu, Z, Yu, Z, Tan, S, Lu, J, Lu, G, Lin, J. | | Deposit date: | 2022-05-01 | | Release date: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Preclinical evaluation of RQ3013, a broad-spectrum mRNA vaccine against SARS-CoV-2 variants.
Sci Bull (Beijing), 68, 2023
|
|
7CQ1
 
 | |
6IHG
 
 | | N terminal domain of Mycobacterium avium complex Lon protease | | Descriptor: | Lon protease | | Authors: | Chen, X.Y, Zhang, S.J, Bi, F.K, Guo, C.Y, Yao, H.W, Lin, D.H. | | Deposit date: | 2018-09-29 | | Release date: | 2019-08-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.397 Å) | | Cite: | Crystal structure of the N domain of Lon protease from Mycobacterium avium complex.
Protein Sci., 28, 2019
|
|
6LQN
 
 | | EBV tegument protein BBRF2 | | Descriptor: | Cytoplasmic envelopment protein 1, NITRATE ION, SULFATE ION | | Authors: | He, H.P, Luo, M, Cao, Y.L, Gao, S. | | Deposit date: | 2020-01-14 | | Release date: | 2020-10-07 | | Last modified: | 2021-04-21 | | Method: | X-RAY DIFFRACTION (1.600022 Å) | | Cite: | Structure of Epstein-Barr virus tegument protein complex BBRF2-BSRF1 reveals its potential role in viral envelopment.
Nat Commun, 11, 2020
|
|
6LQO
 
 | | EBV tegument protein BBRF2/BSRF1 complex | | Descriptor: | ACETATE ION, Cytoplasmic envelopment protein 1, GLYCEROL, ... | | Authors: | He, H.P, Luo, M, Cao, Y.L, Gao, S. | | Deposit date: | 2020-01-14 | | Release date: | 2020-10-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.0911262 Å) | | Cite: | Structure of Epstein-Barr virus tegument protein complex BBRF2-BSRF1 reveals its potential role in viral envelopment.
Nat Commun, 11, 2020
|
|
7DME
 
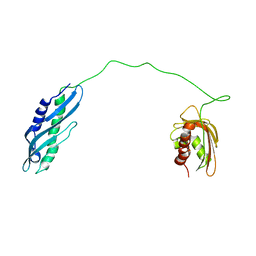 | | Solution structure of human Aha1 | | Descriptor: | Activator of 90 kDa heat shock protein ATPase homolog 1 | | Authors: | Hu, H, Zhou, C, Zhang, N. | | Deposit date: | 2020-12-03 | | Release date: | 2021-04-21 | | Last modified: | 2021-09-22 | | Method: | SOLUTION NMR | | Cite: | Aha1 Exhibits Distinctive Dynamics Behavior and Chaperone-Like Activity.
Molecules, 26, 2021
|
|
7DMD
 
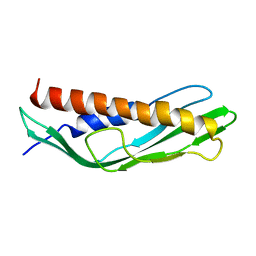 | |
7XVI
 
 | |
