8P72
 
 | | Cryo-EM structure of CAK in complex with inhibitor ICEC0768 | | Descriptor: | CDK-activating kinase assembly factor MAT1, Cyclin-H, Cyclin-dependent kinase 7, ... | | Authors: | Cushing, V.I, Koh, A.F, Feng, J, Jurgaityte, K, Bahl, A.K, Ali, S, Kotecha, A, Greber, B.J. | | Deposit date: | 2023-05-30 | | Release date: | 2024-03-20 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (1.9 Å) | | Cite: | High-resolution cryo-EM of the human CDK-activating kinase for structure-based drug design.
Nat Commun, 15, 2024
|
|
8P70
 
 | | Cryo-EM structure of CAK in complex with inhibitor ICEC0510-S | | Descriptor: | CDK-activating kinase assembly factor MAT1, Cyclin-H, Cyclin-dependent kinase 7, ... | | Authors: | Cushing, V.I, Koh, A.F, Feng, J, Jurgaityte, K, Bahl, A.K, Ali, S, Kotecha, A, Greber, B.J. | | Deposit date: | 2023-05-30 | | Release date: | 2024-03-20 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2 Å) | | Cite: | High-resolution cryo-EM of the human CDK-activating kinase for structure-based drug design.
Nat Commun, 15, 2024
|
|
8P7L
 
 | | Cryo-EM structure of CDK7 subunit of CAK in complex with inhibitor LDC4297 | | Descriptor: | 2-[(3S)-piperidin-3-yl]oxy-8-propan-2-yl-N-[(2-pyrazol-1-ylphenyl)methyl]pyrazolo[1,5-a][1,3,5]triazin-4-amine, CDK-activating kinase assembly factor MAT1, Cyclin-dependent kinase 7 | | Authors: | Cushing, V.I, Koh, A.F, Feng, J, Jurgaityte, K, Bahl, A.K, Ali, S, Kotecha, A, Greber, B.J. | | Deposit date: | 2023-05-30 | | Release date: | 2024-03-20 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.1 Å) | | Cite: | High-resolution cryo-EM of the human CDK-activating kinase for structure-based drug design.
Nat Commun, 15, 2024
|
|
8P79
 
 | | Cryo-EM structure of CAK with averaged inhibitor density | | Descriptor: | CDK-activating kinase assembly factor MAT1, Cyclin-H, Cyclin-dependent kinase 7 | | Authors: | Cushing, V.I, Koh, A.F, Feng, J, Jurgaityte, K, Bahl, A.K, Ali, S, Kotecha, A, Greber, B.J. | | Deposit date: | 2023-05-30 | | Release date: | 2024-03-20 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (1.7 Å) | | Cite: | High-resolution cryo-EM of the human CDK-activating kinase for structure-based drug design.
Nat Commun, 15, 2024
|
|
8P73
 
 | | Cryo-EM structure of CAK in complex with inhibitor ICEC0829 | | Descriptor: | (3R,4R)-4-[[[7-[(phenylmethyl)amino]-3-propan-2-yl-pyrazolo[1,5-a]pyrimidin-5-yl]amino]methyl]piperidine-3,4-diol, (3S,4S)-4-[[[7-[(phenylmethyl)amino]-3-propan-2-yl-pyrazolo[1,5-a]pyrimidin-5-yl]amino]methyl]piperidine-3,4-diol, CDK-activating kinase assembly factor MAT1, ... | | Authors: | Cushing, V.I, Koh, A.F, Feng, J, Jurgaityte, K, Bahl, A.K, Ali, S, Kotecha, A, Greber, B.J. | | Deposit date: | 2023-05-30 | | Release date: | 2024-03-20 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2 Å) | | Cite: | High-resolution cryo-EM of the human CDK-activating kinase for structure-based drug design.
Nat Commun, 15, 2024
|
|
8P6X
 
 | | Cryo-EM structure of CAK in complex with inhibitor BS-194 | | Descriptor: | (2S,3S)-3-{[7-(benzylamino)-3-(1-methylethyl)pyrazolo[1,5-a]pyrimidin-5-yl]amino}butane-1,2,4-triol, CDK-activating kinase assembly factor MAT1, Cyclin-H, ... | | Authors: | Cushing, V.I, Koh, A.F, Feng, J, Jurgaityte, K, Bahl, A.K, Ali, S, Kotecha, A, Greber, B.J. | | Deposit date: | 2023-05-30 | | Release date: | 2024-03-20 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (1.9 Å) | | Cite: | High-resolution cryo-EM of the human CDK-activating kinase for structure-based drug design.
Nat Commun, 15, 2024
|
|
8PLZ
 
 | | Cryo-EM structure of CAK in complex with inhibitor CT7030 | | Descriptor: | (3~{R},4~{R})-4-[[[7-[(2-methoxyphenyl)methylamino]-3-propan-2-yl-pyrazolo[1,5-a]pyrimidin-5-yl]amino]methyl]piperidin-3-ol, CDK-activating kinase assembly factor MAT1, Cyclin-H, ... | | Authors: | Cushing, V.I, Koh, A.F, Feng, J, Jurgaityte, K, Bahl, A.K, Ali, S, Kotecha, A, Greber, B.J. | | Deposit date: | 2023-06-27 | | Release date: | 2024-03-20 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (1.9 Å) | | Cite: | High-resolution cryo-EM of the human CDK-activating kinase for structure-based drug design.
Nat Commun, 15, 2024
|
|
8P74
 
 | | Cryo-EM structure of CAK in complex with inhibitor ICEC0880 (ring-up conformation) | | Descriptor: | (2S,3S)-3-[[7-[(2-bromophenyl)methylamino]-3-propan-2-yl-pyrazolo[1,5-a]pyrimidin-5-yl]amino]butane-1,2,4-triol, CDK-activating kinase assembly factor MAT1, Cyclin-H, ... | | Authors: | Cushing, V.I, Koh, A.F, Feng, J, Jurgaityte, K, Bahl, A.K, Ali, S, Kotecha, A, Greber, B.J. | | Deposit date: | 2023-05-30 | | Release date: | 2024-03-20 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.2 Å) | | Cite: | High-resolution cryo-EM of the human CDK-activating kinase for structure-based drug design.
Nat Commun, 15, 2024
|
|
8P76
 
 | | Cryo-EM structure of CAK in complex with inhibitor ICEC0914 | | Descriptor: | CDK-activating kinase assembly factor MAT1, Cyclin-H, Cyclin-dependent kinase 7, ... | | Authors: | Cushing, V.I, Koh, A.F, Feng, J, Jurgaityte, K, Bahl, A.K, Ali, S, Kotecha, A, Greber, B.J. | | Deposit date: | 2023-05-30 | | Release date: | 2024-03-20 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2 Å) | | Cite: | High-resolution cryo-EM of the human CDK-activating kinase for structure-based drug design.
Nat Commun, 15, 2024
|
|
8QVW
 
 | | Cryo-EM structure of the peptide binding domain of human SRP68/72 | | Descriptor: | Signal recognition particle subunit SRP68, Signal recognition particle subunit SRP72 | | Authors: | Zhong, Y, Feng, J, Koh, A.F, Kotecha, A, Greber, B.J, Ataide, S.F. | | Deposit date: | 2023-10-18 | | Release date: | 2024-02-07 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structure of nPBD of human SRP68/72
To Be Published
|
|
8TZU
 
 | | OC43 S1b domain in complex with WNb 293 and WNb 317 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, ... | | Authors: | Pymm, P, Feng, J, Tham, W.H. | | Deposit date: | 2023-08-27 | | Release date: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | OC43 S1b domain in complex with WNb 293 and WNb 317
To Be Published
|
|
8W7M
 
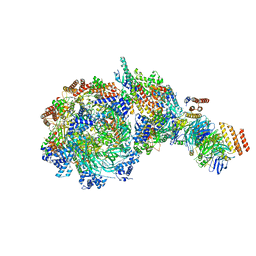 | | Yeast replisome in state V | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Cell division control protein 45, DNA (71-mer), ... | | Authors: | Dang, S, Zhai, Y, Feng, J, Yu, D, Xu, Z. | | Deposit date: | 2023-08-30 | | Release date: | 2023-12-06 | | Method: | ELECTRON MICROSCOPY (4.12 Å) | | Cite: | Synergism between CMG helicase and leading strand DNA polymerase at replication fork.
Nat Commun, 14, 2023
|
|
8W7S
 
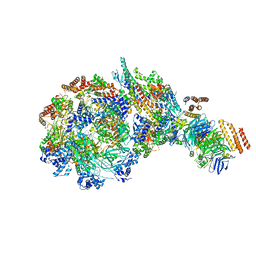 | | Yeast replisome in state IV | | Descriptor: | Cell division control protein 45, DNA (71-mer), DNA polymerase alpha-binding protein, ... | | Authors: | Dang, S, Zhai, Y, Feng, J, Yu, D. | | Deposit date: | 2023-08-31 | | Release date: | 2023-12-20 | | Method: | ELECTRON MICROSCOPY (7.39 Å) | | Cite: | Synergism between CMG helicase and leading strand DNA polymerase at replication fork.
Nat Commun, 14, 2023
|
|
6B0V
 
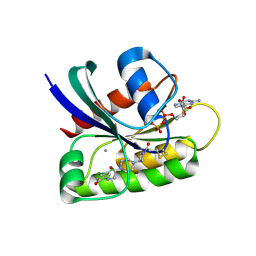 | | Crystal Structure of small molecule ARS-107 covalently bound to K-Ras G12C | | Descriptor: | 1-[3-(4-{[(4,5-dichloro-2-hydroxyphenyl)amino]acetyl}piperazin-1-yl)azetidin-1-yl]propan-1-one, CALCIUM ION, GTPase KRas, ... | | Authors: | Hansen, R, Peters, U, Babbar, A, Chen, Y, Feng, J, Janes, M.R, Li, L.-S, Ren, P, Liu, Y, Zarrinkar, P.P. | | Deposit date: | 2017-09-15 | | Release date: | 2018-05-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | The reactivity-driven biochemical mechanism of covalent KRASG12Cinhibitors.
Nat. Struct. Mol. Biol., 25, 2018
|
|
6B0Y
 
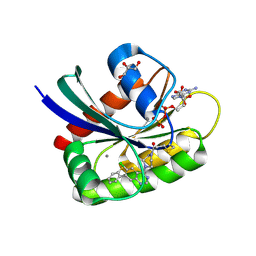 | | Crystal Structure of small molecule ARS-917 covalently bound to K-Ras G12C | | Descriptor: | 1-{4-[6-chloro-7-(2-fluorophenyl)quinazolin-4-yl]piperazin-1-yl}propan-1-one, CALCIUM ION, GLYCEROL, ... | | Authors: | Hansen, R, Peters, U, Babbar, A, Chen, Y, Feng, J, Janes, M.R, Li, L.-S, Ren, P, Liu, Y, Zarrinkar, P.P. | | Deposit date: | 2017-09-15 | | Release date: | 2018-05-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | The reactivity-driven biochemical mechanism of covalent KRASG12Cinhibitors.
Nat. Struct. Mol. Biol., 25, 2018
|
|
8KG8
 
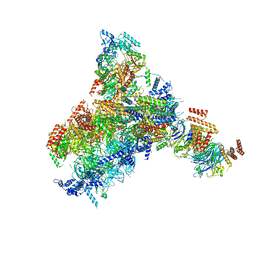 | | Yeast replisome in state II | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Cell division control protein 45, DNA (61-mer), ... | | Authors: | Dang, S, Zhai, Y, Feng, J, Yu, D, Xu, Z. | | Deposit date: | 2023-08-17 | | Release date: | 2023-12-06 | | Method: | ELECTRON MICROSCOPY (4.23 Å) | | Cite: | Synergism between CMG helicase and leading strand DNA polymerase at replication fork.
Nat Commun, 14, 2023
|
|
8KG9
 
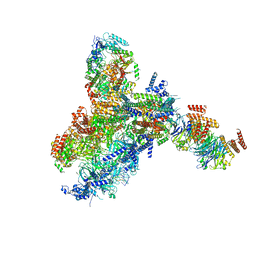 | | Yeast replisome in state III | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Cell division control protein 45, DNA (61-mer), ... | | Authors: | Dang, S, Zhai, Y, Feng, J, Yu, D, Xu, Z. | | Deposit date: | 2023-08-17 | | Release date: | 2023-12-06 | | Method: | ELECTRON MICROSCOPY (4.52 Å) | | Cite: | Synergism between CMG helicase and leading strand DNA polymerase at replication fork.
Nat Commun, 14, 2023
|
|
8KG6
 
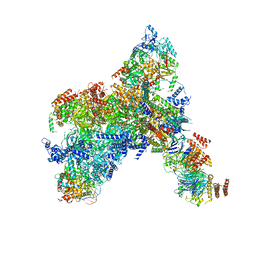 | | Yeast replisome in state I | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Cell division control protein 45, Chromosome segregation in meiosis protein 3, ... | | Authors: | Dang, S, Zhai, Y, Feng, J, Yu, D, Xu, Z. | | Deposit date: | 2023-08-17 | | Release date: | 2023-12-06 | | Method: | ELECTRON MICROSCOPY (3.07 Å) | | Cite: | Synergism between CMG helicase and leading strand DNA polymerase at replication fork.
Nat Commun, 14, 2023
|
|
8WGV
 
 | | BA.2(S375) Spike (S6P)/hACE2 complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Wei, X, Zhang, Z. | | Deposit date: | 2023-09-22 | | Release date: | 2024-02-28 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.92 Å) | | Cite: | The receptor binding domain of SARS-CoV-2 Omicron subvariants targets Siglec-9 to decrease its immunogenicity by preventing macrophage phagocytosis.
Nat.Immunol., 25, 2024
|
|
8WGW
 
 | | Local refinement of RBD-ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Wei, X, Zhang, Z. | | Deposit date: | 2023-09-22 | | Release date: | 2024-02-28 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | The receptor binding domain of SARS-CoV-2 Omicron subvariants targets Siglec-9 to decrease its immunogenicity by preventing macrophage phagocytosis.
Nat.Immunol., 25, 2024
|
|
8XGC
 
 | | Structure of yeast replisome associated with FACT and histone hexamer, Composite map | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Cell division control protein 45, Chromosome segregation in meiosis protein 3, ... | | Authors: | Li, N, Gao, Y, Yu, D, Gao, N, Zhai, Y. | | Deposit date: | 2023-12-15 | | Release date: | 2024-02-14 | | Last modified: | 2024-04-10 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Parental histone transfer caught at the replication fork.
Nature, 627, 2024
|
|
8FY6
 
 | |
8FY7
 
 | | SARS-CoV-2 main protease in complex with covalent inhibitor | | Descriptor: | 3C-like proteinase nsp5, 4-methoxy-N-[(2S)-4-methyl-1-oxo-1-({(2S)-1-[(3S)-2-oxopyrrolidin-3-yl]but-3-en-2-yl}amino)pentan-2-yl]-1H-indole-2-carboxamide | | Authors: | Fried, W, Chen, X.S. | | Deposit date: | 2023-01-25 | | Release date: | 2023-08-30 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Alkyne as a Latent Warhead to Covalently Target SARS-CoV-2 Main Protease.
J.Med.Chem., 66, 2023
|
|
5JI6
 
 | | Potent, Reversible MetAP2 Inhibitors via FBDD | | Descriptor: | 4-(3-methylpyridin-4-yl)-6-(trifluoromethyl)-1H-indazole, MANGANESE (II) ION, Methionine aminopeptidase 2, ... | | Authors: | Dougan, D.R, Lawson, J.D. | | Deposit date: | 2016-04-21 | | Release date: | 2016-05-25 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Discovery of potent, reversible MetAP2 inhibitors via fragment based drug discovery and structure based drug design-Part 1.
Bioorg.Med.Chem.Lett., 26, 2016
|
|
7T9V
 
 | | Crystal structure of hSTING with the agonist, SHR171032 | | Descriptor: | (3S,4S)-4-(3-{5-carbamoyl-2-[(1-ethyl-3-methyl-1H-pyrazole-5-carbonyl)amino]-7-methoxy-1H-benzimidazol-1-yl}propyl)-2-[(1-ethyl-3-methyl-1H-pyrazole-5-carbonyl)amino]-4,5-dihydroimidazo[1,5,4-de][1,4]benzoxazine-8-carboxamide, CALCIUM ION, Stimulator of interferon genes protein | | Authors: | Chowdhury, R, Miller, M. | | Deposit date: | 2021-12-20 | | Release date: | 2022-12-21 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | SHR1032, a novel STING agonist, stimulates anti-tumor immunity and directly induces AML apoptosis.
Sci Rep, 12, 2022
|
|
