4K8D
 
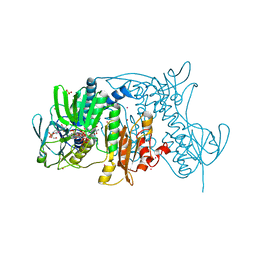 | | Crystal structure of the C558(464)A/C559(465)A double mutant of Tn501 MerA in complex with NADPH and Hg2+ | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, Mercuric reductase, ... | | Authors: | Dong, A, Falkowski, M, Malone, M, Miller, S.M, Pai, E.F. | | Deposit date: | 2013-04-18 | | Release date: | 2013-05-22 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Crystal structure of the C136(42)A/C141(47)A double mutant of Tn501 MerA in complex with NADPH and
Hg2+
to be published
|
|
4K7Z
 
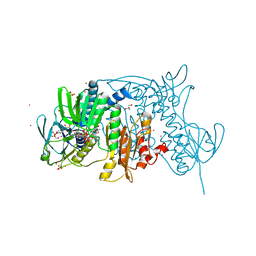 | | Crystal structure of the C136(42)A/C141(47)A double mutant of Tn501 MerA in complex with NADP and Hg2+ | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, MERCURY (II) ION, ... | | Authors: | Dong, A, Falkowaski, M, Malone, M, Miller, S.M, Pai, E.F. | | Deposit date: | 2013-04-17 | | Release date: | 2013-05-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of the C136(42)A/C141(47)A double mutant of Tn501 MerA in complex with NADP and Hg2+
To be Published
|
|
7A8P
 
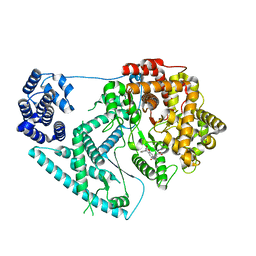 | | Structure of human mitochondrial RNA polymerase in complex with IMT inhibitor. | | Descriptor: | (3~{R})-1-[(2~{R})-2-[4-(2-chloranyl-4-fluoranyl-phenyl)-2-oxidanylidene-chromen-7-yl]oxypropanoyl]piperidine-3-carboxylic acid, DNA-directed RNA polymerase, mitochondrial | | Authors: | Hillen, H.S, Bonekamp, N, Peter, B, Felser, A, Bergbrede, T, Choidas, A, Horn, M, Unger, A, di Lucrezia, R, Atanassov, I, Li, X, Koch, U, Menninger, S, Boros, J, Habenberger, P, Giavalisco, P, Cramer, P, Denzel, M, Nussbaumer, P, Klebl, B, Falkenberg, M, Gustafsson, C.M, Larsson, N.G. | | Deposit date: | 2020-08-30 | | Release date: | 2020-12-30 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Small-molecule inhibitors of human mitochondrial DNA transcription.
Nature, 588, 2020
|
|
1ZX9
 
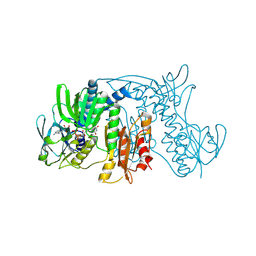 | | Crystal Structure of Tn501 MerA | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Mercuric reductase | | Authors: | Dong, A, Ledwidge, R, Patel, B, Fiedler, D, Falkowski, M, Zelikova, J, Summers, A.O, Pai, E.F, Miller, S.M. | | Deposit date: | 2005-06-07 | | Release date: | 2005-07-05 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | NmerA, the Metal Binding Domain of Mercuric Ion Reductase, Removes Hg(2+) from Proteins, Delivers It to the Catalytic Core, and Protects Cells under Glutathione-Depleted Conditions
Biochemistry, 44, 2005
|
|
1ZK7
 
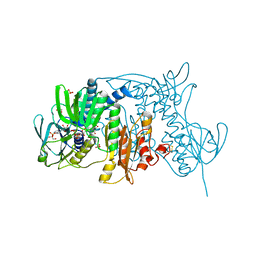 | | Crystal Structure of Tn501 MerA | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, Mercuric reductase, ... | | Authors: | Dong, A, Ledwidge, R, Patel, B, Fiedler, D, Falkowski, M, Zelikova, J, Summers, A.O, Pai, E.F, Miller, S.M. | | Deposit date: | 2005-05-02 | | Release date: | 2005-07-05 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | NmerA, the Metal Binding Domain of Mercuric Ion Reductase, Removes Hg(2+) from Proteins, Delivers It to the Catalytic Core, and Protects Cells under Glutathione-Depleted Conditions
Biochemistry, 44, 2005
|
|
9GGC
 
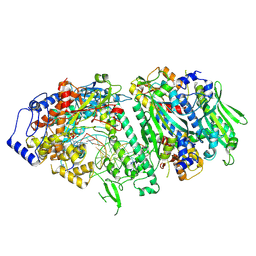 | |
9GGE
 
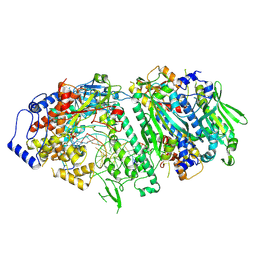 | |
9GGD
 
 | |
9GGF
 
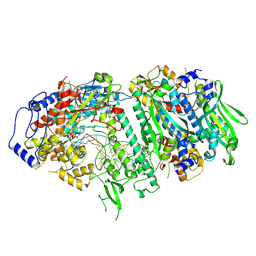 | |
9GGB
 
 | |
4TUU
 
 | | Isolated p110a subunit of PI3Ka provides a platform for structure-based drug design | | Descriptor: | Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Chen, P, Deng, Y.-L, Bergqvist, S, Falk, M, Liu, W, Timofeevski, S. | | Deposit date: | 2014-06-24 | | Release date: | 2014-08-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Engineering of an isolated p110 alpha subunit of PI3K alpha permits crystallization and provides a platform for structure-based drug design.
Protein Sci., 23, 2014
|
|
4TV3
 
 | | Isolated p110a subunit of PI3Ka provides a platform for structure-based drug design | | Descriptor: | 2-amino-8-[trans-4-(2-hydroxyethoxy)cyclohexyl]-6-(6-methoxypyridin-3-yl)-4-methylpyrido[2,3-d]pyrimidin-7(8H)-one, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Chen, P, Deng, Y.-L, Bergqvist, S, Falk, M, Liu, W, Timofeevski, S. | | Deposit date: | 2014-06-25 | | Release date: | 2014-08-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Engineering of an isolated p110 alpha subunit of PI3K alpha permits crystallization and provides a platform for structure-based drug design.
Protein Sci., 23, 2014
|
|
7P0M
 
 | |
7P09
 
 | |
7P0B
 
 | |
6RCI
 
 | | Crystal structure of REXO2 | | Descriptor: | Oligoribonuclease, mitochondrial | | Authors: | Spahr, H, Nicholls, T.J, Larsson, N.G, Gustafsson, C.M. | | Deposit date: | 2019-04-11 | | Release date: | 2019-10-09 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Dinucleotide Degradation by REXO2 Maintains Promoter Specificity in Mammalian Mitochondria.
Mol.Cell, 76, 2019
|
|
6RCN
 
 | | Crystal structure of REXO2-D199A-dAdA | | Descriptor: | DNA (5'-D(P*AP*A)-3'), MAGNESIUM ION, Oligoribonuclease, ... | | Authors: | Spahr, H, Nicholls, T.J, Larsson, N.G, Gustafsson, C.M. | | Deposit date: | 2019-04-11 | | Release date: | 2019-10-09 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Dinucleotide Degradation by REXO2 Maintains Promoter Specificity in Mammalian Mitochondria.
Mol.Cell, 76, 2019
|
|
6RCL
 
 | | Crystal structure of REXO2-D199A-AA | | Descriptor: | MAGNESIUM ION, Oligoribonuclease, mitochondrial, ... | | Authors: | Spahr, H, Nicholls, T.J, Larsson, N.G, Gustafsson, C.M. | | Deposit date: | 2019-04-11 | | Release date: | 2019-10-09 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Dinucleotide Degradation by REXO2 Maintains Promoter Specificity in Mammalian Mitochondria.
Mol.Cell, 76, 2019
|
|
2KT2
 
 | | Structure of NmerA, the N-terminal HMA domain of Tn501 Mercuric Reductase | | Descriptor: | Mercuric reductase | | Authors: | Ledwidge, R, Danacea, F, Dotsch, V, Miller, S.M. | | Deposit date: | 2010-01-17 | | Release date: | 2010-09-22 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | NmerA of Tn501 mercuric ion reductase: structural modulation of the pKa values of the metal binding cysteine thiols.
Biochemistry, 49, 2010
|
|
2KT3
 
 | | Structure of Hg-NmerA, Hg(II) complex of the N-terminal domain of Tn501 Mercuric Reductase | | Descriptor: | MERCURY (II) ION, Mercuric reductase | | Authors: | Miller, S.M, Ledwidge, R, Danacea, F, Dotsch, V. | | Deposit date: | 2010-01-17 | | Release date: | 2010-09-22 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | NmerA of Tn501 mercuric ion reductase: structural modulation of the pKa values of the metal binding cysteine thiols.
Biochemistry, 49, 2010
|
|
7PZR
 
 | | Cryo-EM structure of POLRMT in free form. | | Descriptor: | DNA-directed RNA polymerase, mitochondrial | | Authors: | Das, H, Hallberg, B.M. | | Deposit date: | 2021-10-13 | | Release date: | 2022-11-16 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Non-coding 7S RNA inhibits transcription via mitochondrial RNA polymerase dimerization.
Cell, 185, 2022
|
|
7PZP
 
 | |
7ZC4
 
 | | Cryo-EM structure of POLRMT mutant. | | Descriptor: | DNA-directed RNA polymerase, mitochondrial | | Authors: | Das, H, Hallberg, B.M. | | Deposit date: | 2022-03-25 | | Release date: | 2023-02-01 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.24 Å) | | Cite: | Non-coding 7S RNA inhibits transcription via mitochondrial RNA polymerase dimerization.
Cell, 185, 2022
|
|
