3ITK
 
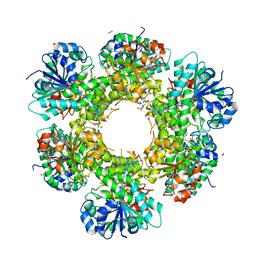 | | Crystal structure of human UDP-glucose dehydrogenase Thr131Ala, apo form. | | Descriptor: | 1,2-ETHANEDIOL, TETRAETHYLENE GLYCOL, UDP-glucose 6-dehydrogenase | | Authors: | Chaikuad, A, Egger, S, Yue, W.W, Sethi, R, Filippakopoulos, P, Muniz, J.R.C, von Delft, F, Bountra, C, Arrowsmith, C.H, Weigelt, J, Edwards, A.M, Kavanagh, K.L, Nidetzky, B, Oppermann, U, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-08-28 | | Release date: | 2009-09-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure and mechanism of human UDP-glucose 6-dehydrogenase.
J.Biol.Chem., 286, 2011
|
|
3KHU
 
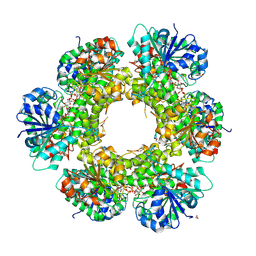 | | Crystal structure of human UDP-glucose dehydrogenase Glu161Gln, in complex with thiohemiacetal intermediate | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, UDP-glucose 6-dehydrogenase, ... | | Authors: | Chaikuad, A, Egger, S, Yue, W.W, Guo, K, Sethi, R, Filippakopoulos, P, Muniz, J.R.C, von Delft, F, Bountra, C, Arrowsmith, C.H, Weigelt, J, Edwards, A.M, Kavanagh, K.L, Nidetzky, B, Oppermann, U, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-10-30 | | Release date: | 2009-11-17 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and Kinetic Evidence That Catalytic Reaction of Human UDP-glucose 6-Dehydrogenase Involves Covalent Thiohemiacetal and Thioester Enzyme Intermediates.
J.Biol.Chem., 287, 2012
|
|
2QG4
 
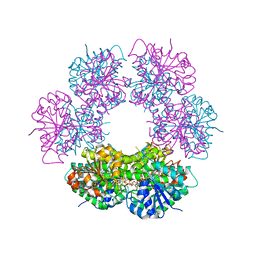 | | Crystal structure of human UDP-glucose dehydrogenase product complex with UDP-glucuronate | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Kavanagh, K.L, Guo, K, Bunkoczi, G, Savitsky, P, Pilka, E, Bhatia, C, Niesen, F, Smee, C, Berridge, G, von Delft, F, Weigelt, J, Arrowsmith, C.H, Sundstrom, M, Edwards, A, Oppermann, U, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-06-28 | | Release date: | 2007-07-10 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure and mechanism of human UDP-glucose 6-dehydrogenase.
J.Biol.Chem., 286, 2011
|
|
2Q3E
 
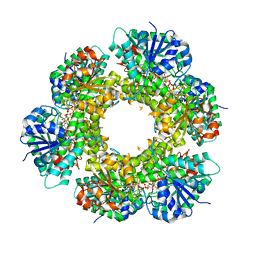 | | Structure of human UDP-glucose dehydrogenase complexed with NADH and UDP-glucose | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, CHLORIDE ION, UDP-glucose 6-dehydrogenase, ... | | Authors: | Kavanagh, K.L, Guo, K, Bunkoczi, G, Savitsky, P, Pilka, E, Bhatia, C, Smee, C, Berridge, G, von Delft, F, Wiegelt, J, Arrowsmith, C, Sundstrom, M, Edwards, A, Oppermann, U, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-05-30 | | Release date: | 2007-07-03 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure and mechanism of human UDP-glucose 6-dehydrogenase.
J.Biol.Chem., 286, 2011
|
|
4LU4
 
 | |
4WGJ
 
 | |
4JFN
 
 | | Crystal structure of the N-terminal, growth factor-like domain of the amyloid precursor protein bound to copper | | Descriptor: | Amyloid beta A4 protein, COPPER (II) ION, GLYCEROL | | Authors: | Wild, K, Baumkotter, F, Kins, S. | | Deposit date: | 2013-02-28 | | Release date: | 2014-07-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Amyloid precursor protein dimerization and synaptogenic function depend on copper binding to the growth factor-like domain
J.Neurosci., 34, 2014
|
|
6HH0
 
 | |
5NQH
 
 | | Structure of the human Fe65-PTB2 homodimer | | Descriptor: | Amyloid beta A4 precursor protein-binding family B member 1, GLYCEROL, SULFATE ION | | Authors: | Feilen, L.P, Haubrich, K, Sinning, I, Konietzko, U, Kins, S, Simon, B, Wild, K. | | Deposit date: | 2017-04-20 | | Release date: | 2017-05-03 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Fe65-PTB2 Dimerization Mimics Fe65-APP Interaction.
Front Mol Neurosci, 10, 2017
|
|
4NPS
 
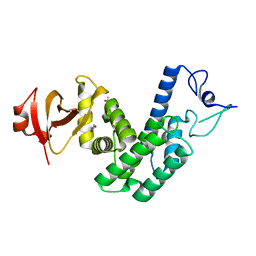 | | Crystal Structure of Bep1 protein (VirB-translocated Bartonella effector protein) from Bartonella clarridgeiae | | Descriptor: | ACETATE ION, Bartonella effector protein (Bep) substrate of VirB T4SS | | Authors: | Dranow, D.M, Abendroth, J, Edwards, T.E, Lorimer, D, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2013-11-22 | | Release date: | 2014-10-08 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Evolutionary Diversification of Host-Targeted Bartonella Effectors Proteins Derived from a Conserved FicTA Toxin-Antitoxin Module.
Microorganisms, 9, 2021
|
|
4PY3
 
 | |
4N67
 
 | |
4XI8
 
 | |
4M16
 
 | |
