6DMX
 
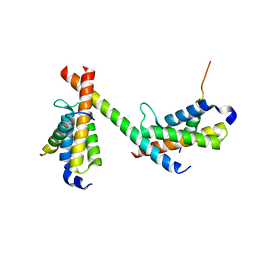 | | HBZ56 in complex with KIX and c-Myb | | 分子名称: | BZIP factor, CREB-binding protein, Transcriptional activator Myb | | 著者 | Yang, K, Wright, P.E, Stanfield, R.L. | | 登録日 | 2018-06-05 | | 公開日 | 2018-09-19 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Structural basis for cooperative regulation of KIX-mediated transcription pathways by the HTLV-1 HBZ activation domain.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6DNQ
 
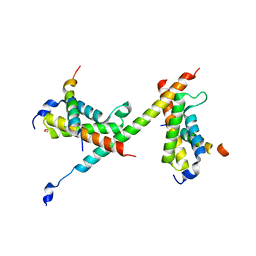 | | HBZ77 in complex with KIX and c-Myb | | 分子名称: | 1,2-ETHANEDIOL, BZIP factor, CREB-binding protein, ... | | 著者 | Yang, K, Wright, P.E, Stanfield, R.L. | | 登録日 | 2018-06-07 | | 公開日 | 2018-09-19 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.35 Å) | | 主引用文献 | Structural basis for cooperative regulation of KIX-mediated transcription pathways by the HTLV-1 HBZ activation domain.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
1AI1
 
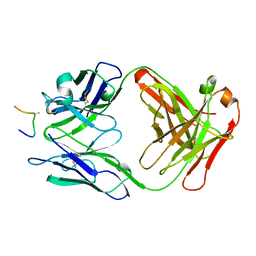 | | HIV-1 V3 LOOP MIMIC | | 分子名称: | AIB142, IGG1-KAPPA 59.1 FAB (HEAVY CHAIN), IGG1-KAPPA 59.1 FAB (LIGHT CHAIN) | | 著者 | Ghiara, J.B, Wilson, I.A. | | 登録日 | 1996-11-06 | | 公開日 | 1997-05-15 | | 最終更新日 | 2023-08-02 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Structure-based design of a constrained peptide mimic of the HIV-1 V3 loop neutralization site.
J.Mol.Biol., 266, 1997
|
|
5W3Q
 
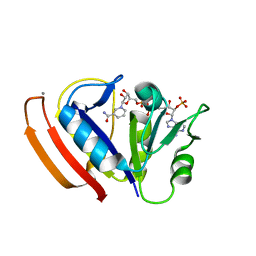 | | L28F E.coli DHFR in complex with NADPH | | 分子名称: | CALCIUM ION, Dihydrofolate reductase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | 著者 | Oyen, D, Wright, P.E, Wilson, I.A. | | 登録日 | 2017-06-08 | | 公開日 | 2017-08-09 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.401 Å) | | 主引用文献 | Defining the Structural Basis for Allosteric Product Release from E. coli Dihydrofolate Reductase Using NMR Relaxation Dispersion.
J. Am. Chem. Soc., 139, 2017
|
|
4M6L
 
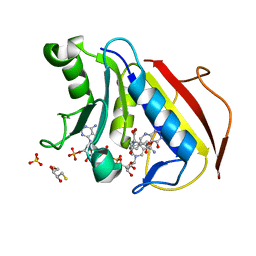 | | Crystal structure of human dihydrofolate reductase (DHFR) bound to NADP+ and 5,10-dideazatetrahydrofolic acid | | 分子名称: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, Dihydrofolate reductase, N-(4-{2-[(6S)-2-amino-4-oxo-1,4,5,6,7,8-hexahydropyrido[2,3-d]pyrimidin-6-yl]ethyl}benzoyl)-L-glutamic acid, ... | | 著者 | Bhabha, G, Ekiert, D.C, Wright, P.E, Wilson, I.A. | | 登録日 | 2013-08-09 | | 公開日 | 2013-09-25 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Divergent evolution of protein conformational dynamics in dihydrofolate reductase.
Nat.Struct.Mol.Biol., 20, 2013
|
|
4M6J
 
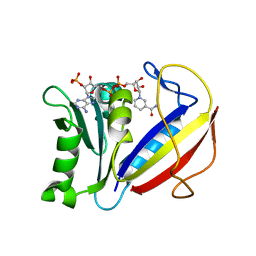 | | Crystal structure of human dihydrofolate reductase (DHFR) bound to NADPH | | 分子名称: | Dihydrofolate reductase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | 著者 | Bhabha, G, Ekiert, D.C, Wright, P.E, Wilson, I.A. | | 登録日 | 2013-08-09 | | 公開日 | 2013-09-25 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.201 Å) | | 主引用文献 | Divergent evolution of protein conformational dynamics in dihydrofolate reductase.
Nat.Struct.Mol.Biol., 20, 2013
|
|
4M6K
 
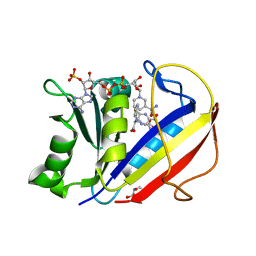 | | Crystal structure of human dihydrofolate reductase (DHFR) bound to NADP+ and folate | | 分子名称: | Dihydrofolate reductase, FOLIC ACID, GLYCEROL, ... | | 著者 | Bhabha, G, Ekiert, D.C, Wright, P.E, Wilson, I.A. | | 登録日 | 2013-08-09 | | 公開日 | 2013-09-25 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.396 Å) | | 主引用文献 | Divergent evolution of protein conformational dynamics in dihydrofolate reductase.
Nat.Struct.Mol.Biol., 20, 2013
|
|
3QL3
 
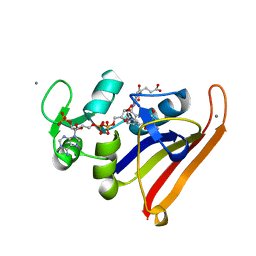 | | Re-refined coordinates for PDB entry 1RX2 | | 分子名称: | Dihydrofolate reductase, FOLIC ACID, MANGANESE (II) ION, ... | | 著者 | Bhabha, G, Ekiert, D.C, Wright, P.E, Wilson, I.A. | | 登録日 | 2011-02-02 | | 公開日 | 2011-04-27 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | A dynamic knockout reveals that conformational fluctuations influence the chemical step of enzyme catalysis.
Science, 332, 2011
|
|
3QL0
 
 | | Crystal structure of N23PP/S148A mutant of E. coli dihydrofolate reductase | | 分子名称: | Dihydrofolate reductase, FOLIC ACID, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | 著者 | Bhabha, G, Ekiert, D.C, Wright, P.E, Wilson, I.A. | | 登録日 | 2011-02-02 | | 公開日 | 2011-04-20 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | A dynamic knockout reveals that conformational fluctuations influence the chemical step of enzyme catalysis.
Science, 332, 2011
|
|
1RCY
 
 | | RUSTICYANIN (RC) FROM THIOBACILLUS FERROOXIDANS | | 分子名称: | COPPER (II) ION, RUSTICYANIN | | 著者 | Walter, R.L, Friedman, A.M, Ealick, S.E, Blake II, R.C, Proctor, P, Shoham, M. | | 登録日 | 1996-04-10 | | 公開日 | 1997-05-15 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Multiple wavelength anomalous diffraction (MAD) crystal structure of rusticyanin: a highly oxidizing cupredoxin with extreme acid stability.
J.Mol.Biol., 263, 1996
|
|
1Z65
 
 | | Mouse Doppel 1-30 peptide | | 分子名称: | Prion-like protein doppel | | 著者 | Papadopoulos, E. | | 登録日 | 2005-03-21 | | 公開日 | 2006-02-07 | | 最終更新日 | 2022-03-02 | | 実験手法 | SOLUTION NMR | | 主引用文献 | NMR Solution Structure of the Peptide Fragment 1-30, Derived from Unprocessed Mouse Doppel Protein, in DHPC Micelles
Biochemistry, 45, 2006
|
|
2JOX
 
 | | Embryonic Neural Inducing Factor Churchill is not a DNA-Binding Zinc Finger Protein: Solution Structure Reveals a Solvent-Exposed beta-Sheet and Zinc Binuclear Cluster | | 分子名称: | Churchill protein, ZINC ION | | 著者 | Lee, B.M, Buck-Koehntop, B.A, Martinez-Yamout, M.A, Gottesfeld, J.M, Dyson, H, Wright, P.E. | | 登録日 | 2007-04-07 | | 公開日 | 2007-08-21 | | 最終更新日 | 2024-05-08 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Embryonic Neural Inducing Factor Churchill Is not a DNA-binding Zinc Finger Protein: Solution Structure Reveals a Solvent-exposed beta-Sheet and Zinc Binuclear Cluster
J.Mol.Biol., 371, 2007
|
|
2KJE
 
 | | NMR structure of CBP TAZ2 and adenoviral E1A complex | | 分子名称: | CREB-binding protein, Early E1A 32 kDa protein, ZINC ION | | 著者 | Ferreon, J.C, Martinez-Yamout, M, Dyson, H, Wright, P.E. | | 登録日 | 2009-05-27 | | 公開日 | 2009-09-15 | | 最終更新日 | 2022-03-16 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structural basis for subversion of cellular control mechanisms by the adenoviral E1A oncoprotein.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
2LT7
 
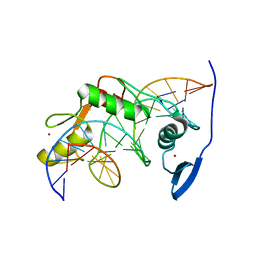 | | Solution NMR structure of Kaiso zinc finger DNA binding domain in complex with Kaiso binding site DNA | | 分子名称: | DNA (5'-D(*CP*GP*TP*TP*AP*TP*TP*GP*GP*CP*AP*GP*GP*AP*AP*GP*CP*AP*C)-3'), DNA (5'-D(*GP*TP*GP*CP*TP*TP*CP*CP*TP*GP*CP*CP*AP*AP*TP*AP*AP*CP*G)-3'), Transcriptional regulator Kaiso, ... | | 著者 | Buck-Koehntop, B.A, Stanfield, R.L, Ekiert, D.C, Martinez-Yamout, M.A, Dyson, H, Wilson, I.A, Wright, P.E. | | 登録日 | 2012-05-15 | | 公開日 | 2012-09-05 | | 最終更新日 | 2024-05-01 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Molecular basis for recognition of methylated and specific DNA sequences by the zinc finger protein Kaiso.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
2MKN
 
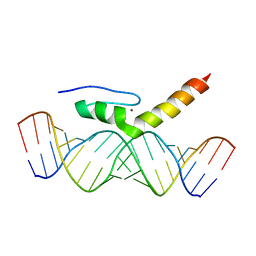 | | Structural Characterization of Interactions between the Double-Stranded RNA-Binding Zinc Finger Protein JAZ and dsRNA | | 分子名称: | RNA (5'-R(*GP*CP*CP*GP*UP*GP*GP*UP*CP*UP*GP*GP*UP*GP*GP*CP*CP*GP*G)-3'), RNA (5'-R(P*CP*CP*GP*GP*CP*CP*AP*CP*CP*AP*GP*AP*CP*CP*AP*CP*GP*GP*C)-3'), ZINC ION, ... | | 著者 | Wright, P, Dyson, J, Burge, R, Martinez-Yamout, M. | | 登録日 | 2014-02-10 | | 公開日 | 2014-03-19 | | 最終更新日 | 2024-05-01 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structural Characterization of Interactions between the Double-Stranded RNA-Binding Zinc Finger Protein JAZ and Nucleic Acids.
Biochemistry, 53, 2014
|
|
2MKD
 
 | | Human JAZ ZF3 Residues 168-227 | | 分子名称: | ZINC ION, Zinc finger protein 346 | | 著者 | Wright, P, Dyson, J, Burge, R, Martinez-Yamout, M. | | 登録日 | 2014-02-05 | | 公開日 | 2014-03-19 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structural Characterization of Interactions between the Double-Stranded RNA-Binding Zinc Finger Protein JAZ and Nucleic Acids.
Biochemistry, 53, 2014
|
|
2NC5
 
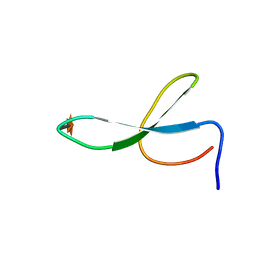 | | Solution Structure of N-Xylosylated Pin1 WW Domain | | 分子名称: | Pin1 WW Domain, beta-D-xylopyranose | | 著者 | Hsu, C, Park, S, Mortenson, D.E, Foley, B, Wang, X, Woods, R.J, Case, D.A, Powers, E.T, Wong, C, Dyson, H, Kelly, J.W. | | 登録日 | 2016-03-20 | | 公開日 | 2016-06-08 | | 最終更新日 | 2020-07-29 | | 実験手法 | SOLUTION NMR | | 主引用文献 | The Dependence of Carbohydrate-Aromatic Interaction Strengths on the Structure of the Carbohydrate.
J.Am.Chem.Soc., 138, 2016
|
|
2NC4
 
 | | Solution Structure of N-Galactosylated Pin1 WW Domain | | 分子名称: | Pin1 WW Domain, beta-D-galactopyranose | | 著者 | Hsu, C, Park, S, Mortenson, D.E, Foley, B, Wang, X, Woods, R.J, Case, D.A, Powers, E.T, Wong, C, Dyson, H, Kelly, J.W. | | 登録日 | 2016-03-20 | | 公開日 | 2016-06-08 | | 最終更新日 | 2023-06-14 | | 実験手法 | SOLUTION NMR | | 主引用文献 | The Dependence of Carbohydrate-Aromatic Interaction Strengths on the Structure of the Carbohydrate.
J.Am.Chem.Soc., 138, 2016
|
|
2NC6
 
 | | Solution Structure of N-L-idosylated Pin1 WW Domain | | 分子名称: | Pin1 WW Domain, beta-L-idopyranose | | 著者 | Hsu, C, Park, S, Mortenson, D.E, Foley, B, Wang, X, Woods, R.J, Case, D.A, Powers, E.T, Wong, C, Dyson, H, Kelly, J.W. | | 登録日 | 2016-03-20 | | 公開日 | 2016-06-08 | | 最終更新日 | 2023-06-14 | | 実験手法 | SOLUTION NMR | | 主引用文献 | The Dependence of Carbohydrate-Aromatic Interaction Strengths on the Structure of the Carbohydrate.
J.Am.Chem.Soc., 138, 2016
|
|
2NC3
 
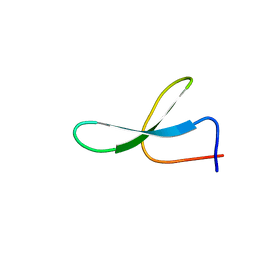 | | Solution Structure of N-Allosylated Pin1 WW Domain | | 分子名称: | Pin1 WW Domain, beta-D-allopyranose | | 著者 | Hsu, C, Park, S, Mortenson, D.E, Foley, B, Wang, X, Woods, R.J, Case, D.A, Powers, E.T, Wong, C, Dyson, H, Kelly, J.W. | | 登録日 | 2016-03-20 | | 公開日 | 2016-06-08 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | The Dependence of Carbohydrate-Aromatic Interaction Strengths on the Structure of the Carbohydrate.
J.Am.Chem.Soc., 138, 2016
|
|
7D9L
 
 | |
1A3Z
 
 | | REDUCED RUSTICYANIN AT 1.9 ANGSTROMS | | 分子名称: | COPPER (I) ION, RUSTICYANIN | | 著者 | Zhao, D, Shoham, M. | | 登録日 | 1998-01-27 | | 公開日 | 1998-07-29 | | 最終更新日 | 2023-08-02 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Rusticyanin: Extremes in acid stability and redox potential explained by the crystal structure.
Biophys.J., 74, 1998
|
|
1A8Z
 
 | | STRUCTURE DETERMINATION OF A 16.8KDA COPPER PROTEIN RUSTICYANIN AT 2.1A RESOLUTION USING ANOMALOUS SCATTERING DATA WITH DIRECT METHODS | | 分子名称: | COPPER (I) ION, RUSTICYANIN | | 著者 | Harvey, I, Hao, Q, Duke, E.M.H, Ingledew, W.J, Hasnain, S.S. | | 登録日 | 1998-03-30 | | 公開日 | 1998-06-17 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structure determination of a 16.8 kDa copper protein at 2.1 A resolution using anomalous scattering data with direct methods.
Acta Crystallogr.,Sect.D, 54, 1998
|
|
2KBU
 
 | |
