2VJU
 
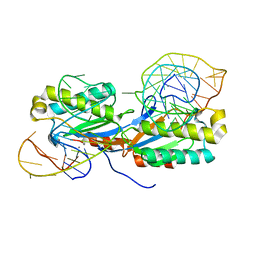 | | Crystal structure of the IS608 transposase in complex with the complete Right end 35-mer DNA and manganese | | 分子名称: | MANGANESE (II) ION, RIGHT END 35-MER, TRANSPOSASE ORFA | | 著者 | Barabas, O, Ronning, D.R, Guynet, C, Hickman, A.B, Ton-Hoang, B, Chandler, M, Dyda, F. | | 登録日 | 2007-12-13 | | 公開日 | 2008-02-19 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Mechanism of is200/is605 Family DNA Transposases: Activation and Transposon-Directed Target Site Selection.
Cell(Cambridge,Mass.), 132, 2008
|
|
2V0X
 
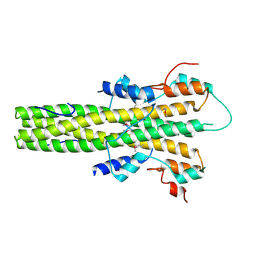 | | The dimerization domain of LAP2alpha | | 分子名称: | LAMINA-ASSOCIATED POLYPEPTIDE 2 ISOFORMS ALPHA/ZETA | | 著者 | Bradley, C.M, Jones, S, Huang, Y, Suzuki, Y, Kvaratskhelia, M, Hickman, A.B, Craigie, R, Dyda, F. | | 登録日 | 2007-05-20 | | 公開日 | 2007-06-26 | | 最終更新日 | 2019-05-15 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Structural Basis for Dimerization of Lap2Alpha, a Component of the Nuclear Lamina.
Structure, 15, 2007
|
|
2VIH
 
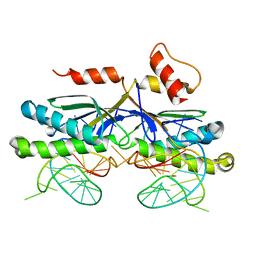 | | CRYSTAL STRUCTURE OF THE IS608 TRANSPOSASE IN COMPLEX WITH Left END 26-MER DNA | | 分子名称: | 5'-D(*AP*AP*AP*GP*CP*CP*CP*CP*TP*AP *GP*CP*TP*TP*TP*TP*AP*GP*CP*TP*AP*TP*GP*GP*GP*G)-3', TRANSPOSASE ORFA | | 著者 | Barabas, O, Ronning, D.R, Guynet, C, Hickman, A.B, Ton-Hoang, B, Chandler, M, Dyda, F. | | 登録日 | 2007-12-04 | | 公開日 | 2008-02-19 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Mechanism of is200/is605 Family DNA Transposases: Activation and Transposon-Directed Target Site Selection.
Cell(Cambridge,Mass.), 132, 2008
|
|
2VIC
 
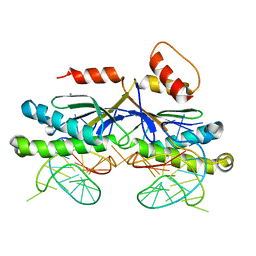 | | CRYSTAL STRUCTURE OF THE ISHP608 TRANSPOSASE IN COMPLEX with Left end 26- mer DNA and manganese | | 分子名称: | 5'-D(*AP*AP*AP*GP*CP*CP*CP*CP*TP*AP *GP*CP*TP*TP*TP*TP*AP*GP*CP*TP*AP*TP*GP*GP*GP*G)-3', MANGANESE (II) ION, TRANSPOSASE ORFA | | 著者 | Barabas, O, Ronning, D.R, Guynet, C, Hickman, A.B, Ton-Hoang, B, Chandler, M, Dyda, F. | | 登録日 | 2007-11-29 | | 公開日 | 2008-02-19 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2.35 Å) | | 主引用文献 | Mechanism of is200/is605 Family DNA Transposases: Activation and Transposon-Directed Target Site Selection.
Cell(Cambridge,Mass.), 132, 2008
|
|
2VHG
 
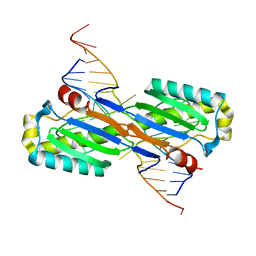 | | Crystal Structure of the ISHp608 Transposase in Complex with Right End 31-mer DNA | | 分子名称: | MANGANESE (II) ION, RIGHT END 31-MER, TRANSPOSASE ORFA | | 著者 | Barabas, O, Ronning, D.R, Guynet, C, Hickman, A.B, Ton-Hoang, B, Chandler, M, Dyda, F. | | 登録日 | 2007-11-21 | | 公開日 | 2008-02-19 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Mechanism of is200/is605 Family DNA Transposases: Activation and Transposon-Directed Target Site Selection.
Cell(Cambridge,Mass.), 132, 2008
|
|
2X2E
 
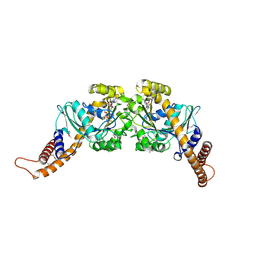 | | Dynamin GTPase dimer, long axis form | | 分子名称: | DYNAMIN-1, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | 著者 | Chappie, J.S, Acharya, S, Leonard, M, Schmid, S.L, Dyda, F. | | 登録日 | 2010-01-12 | | 公開日 | 2010-04-28 | | 最終更新日 | 2019-05-08 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | G Domain Dimerization Controls Dynamin'S Assembly-Stimulated Gtpase Activity.
Nature, 465, 2010
|
|
1AIH
 
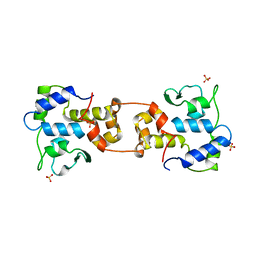 | | CATALYTIC DOMAIN OF BACTERIOPHAGE HP1 INTEGRASE | | 分子名称: | HP1 INTEGRASE, MAGNESIUM ION, SULFATE ION | | 著者 | Hickman, A.B, Waninger, S, Scocca, J.J, Dyda, F. | | 登録日 | 1997-04-17 | | 公開日 | 1997-08-20 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Molecular organization in site-specific recombination: the catalytic domain of bacteriophage HP1 integrase at 2.7 A resolution.
Cell(Cambridge,Mass.), 89, 1997
|
|
1A30
 
 | | HIV-1 PROTEASE COMPLEXED WITH A TRIPEPTIDE INHIBITOR | | 分子名称: | HIV-1 PROTEASE, TRIPEPTIDE GLU-ASP-LEU | | 著者 | Louis, J.M, Dyda, F, Nashed, N.T, Kimmel, A.R, Davies, D.R. | | 登録日 | 1998-01-27 | | 公開日 | 1998-04-29 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Hydrophilic peptides derived from the transframe region of Gag-Pol inhibit the HIV-1 protease.
Biochemistry, 37, 1998
|
|
1B6B
 
 | |
1CJW
 
 | | SEROTONIN N-ACETYLTRANSFERASE COMPLEXED WITH A BISUBSTRATE ANALOG | | 分子名称: | COA-S-ACETYL TRYPTAMINE, PROTEIN (SEROTONIN N-ACETYLTRANSFERASE) | | 著者 | Hickman, A.B, Namboodiri, M.A.A, Klein, D.C, Dyda, F. | | 登録日 | 1999-04-19 | | 公開日 | 1999-05-06 | | 最終更新日 | 2023-08-09 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | The structural basis of ordered substrate binding by serotonin N-acetyltransferase: enzyme complex at 1.8 A resolution with a bisubstrate analog.
Cell(Cambridge,Mass.), 97, 1999
|
|
1PVD
 
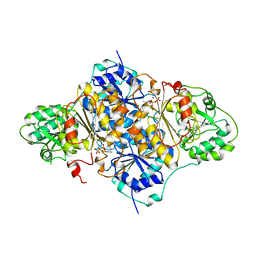 | |
2ITG
 
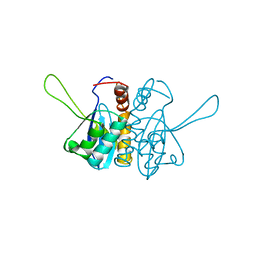 | | CATALYTIC DOMAIN OF HIV-1 INTEGRASE: ORDERED ACTIVE SITE IN THE F185H CONSTRUCT | | 分子名称: | HUMAN IMMUNODEFICIENCY VIRUS-1 INTEGRASE | | 著者 | Bujacz, G, Alexandratos, J, Wlodawer, A, Zhou-Liu, Q, Clement-Mella, C. | | 登録日 | 1996-09-13 | | 公開日 | 1997-03-12 | | 最終更新日 | 2023-08-09 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | The catalytic domain of human immunodeficiency virus integrase: ordered active site in the F185H mutant.
FEBS Lett., 398, 1996
|
|
6NSN
 
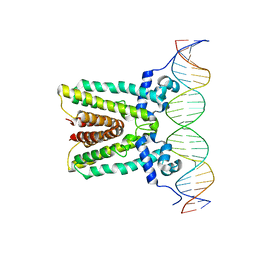 | |
6NSM
 
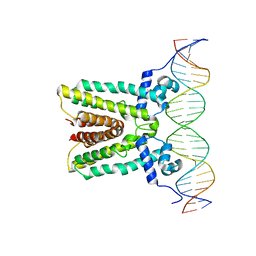 | |
6NSR
 
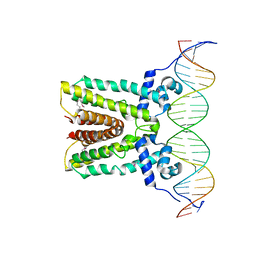 | |
6FI8
 
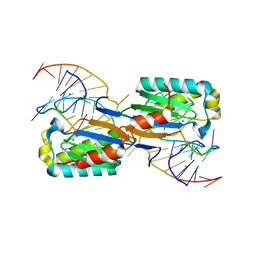 | |
1AKN
 
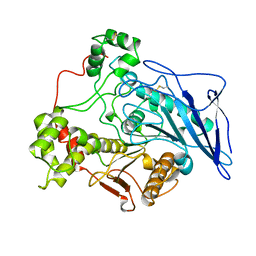 | | STRUCTURE OF BILE-SALT ACTIVATED LIPASE | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, BILE-SALT ACTIVATED LIPASE | | 著者 | Wang, X, Zhang, X. | | 登録日 | 1997-05-23 | | 公開日 | 1998-05-27 | | 最終更新日 | 2023-08-02 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | The crystal structure of bovine bile salt activated lipase: insights into the bile salt activation mechanism.
Structure, 5, 1997
|
|
1AQL
 
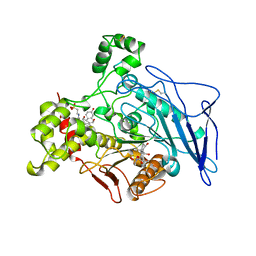 | |
1IKP
 
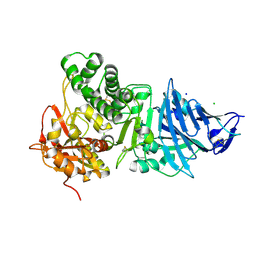 | | Pseudomonas Aeruginosa Exotoxin A, P201Q, W281A mutant | | 分子名称: | CHLORIDE ION, EXOTOXIN A, SODIUM ION | | 著者 | McKay, D.B, Wedekind, J.E, Trame, C.B. | | 登録日 | 2001-05-04 | | 公開日 | 2001-12-12 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.45 Å) | | 主引用文献 | Refined Crystallographic Structure of Pseudomonas aeruginosa Exotoxin A
and its Implications for the Molecular Mechanism of Toxicity
J.Mol.Biol., 314, 2001
|
|
1IKQ
 
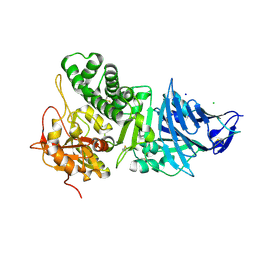 | | Pseudomonas Aeruginosa Exotoxin A, wild type | | 分子名称: | CHLORIDE ION, EXOTOXIN A, SODIUM ION | | 著者 | McKay, D.B, Wedekind, J.E, Trame, C.B. | | 登録日 | 2001-05-04 | | 公開日 | 2001-12-12 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.62 Å) | | 主引用文献 | Refined Crystallographic Structure of Pseudomonas aeruginosa Exotoxin A
and its Implications for the Molecular Mechanism of Toxicity
J.Mol.Biol., 314, 2001
|
|
1Q10
 
 | |
1BI4
 
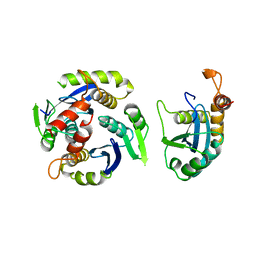 | | CATALYTIC DOMAIN OF HIV-1 INTEGRASE | | 分子名称: | INTEGRASE | | 著者 | Maignan, S, Guilloteau, J.P, Zhou-Liu, Q, Clement-Mella, C, Mikol, V. | | 登録日 | 1998-06-22 | | 公開日 | 1998-11-04 | | 最終更新日 | 2023-08-02 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Crystal structures of the catalytic domain of HIV-1 integrase free and complexed with its metal cofactor: high level of similarity of the active site with other viral integrases.
J.Mol.Biol., 282, 1998
|
|
1BHL
 
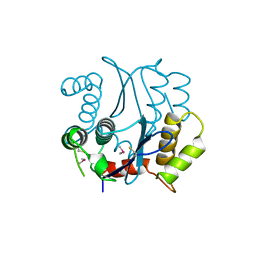 | | CACODYLATED CATALYTIC DOMAIN OF HIV-1 INTEGRASE | | 分子名称: | HIV-1 INTEGRASE | | 著者 | Maignan, S, Guilloteau, J.P, Zhou-Liu, Q, Clement-Mella, C, Mikol, V. | | 登録日 | 1998-06-10 | | 公開日 | 1998-10-14 | | 最終更新日 | 2023-08-02 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Crystal structures of the catalytic domain of HIV-1 integrase free and complexed with its metal cofactor: high level of similarity of the active site with other viral integrases.
J.Mol.Biol., 282, 1998
|
|
1BL3
 
 | | CATALYTIC DOMAIN OF HIV-1 INTEGRASE | | 分子名称: | INTEGRASE, MAGNESIUM ION | | 著者 | Maignan, S, Guilloteau, J.P, Zhou-Liu, Q, Clement-Mella, C, Mikol, V. | | 登録日 | 1998-07-23 | | 公開日 | 1998-09-30 | | 最終更新日 | 2023-08-02 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Crystal structures of the catalytic domain of HIV-1 integrase free and complexed with its metal cofactor: high level of similarity of the active site with other viral integrases.
J.Mol.Biol., 282, 1998
|
|
