6N50
 
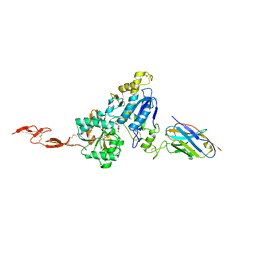 | | Metabotropic Glutamate Receptor 5 Extracellular Domain in Complex with Nb43 and L-quisqualic acid | | Descriptor: | (S)-2-AMINO-3-(3,5-DIOXO-[1,2,4]OXADIAZOLIDIN-2-YL)-PROPIONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, Metabotropic glutamate receptor 5, ... | | Authors: | Koehl, A, Hu, H, Feng, D, Sun, B, Chu, M, Weis, W.I, Skiniotis, G, Mathiesen, J.M, Kobilka, B.K. | | Deposit date: | 2018-11-20 | | Release date: | 2019-01-23 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.751 Å) | | Cite: | Structural insights into the activation of metabotropic glutamate receptors.
Nature, 566, 2019
|
|
6N4Y
 
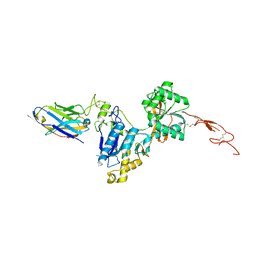 | | Metabotropic Glutamate Receptor 5 Extracellular Domain with Nb43 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Metabotropic glutamate receptor 5, ... | | Authors: | Koehl, A, Hu, H, Feng, D, Sun, B, Chu, M, Weis, W.I, Mathiesen, J.M, Skiniotis, G, Kobilka, B.K. | | Deposit date: | 2018-11-20 | | Release date: | 2019-01-23 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.262 Å) | | Cite: | Structural insights into the activation of metabotropic glutamate receptors.
Nature, 566, 2019
|
|
6N51
 
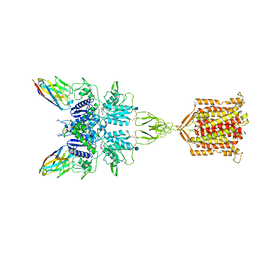 | | Metabotropic Glutamate Receptor 5 bound to L-quisqualate and Nb43 | | Descriptor: | (S)-2-AMINO-3-(3,5-DIOXO-[1,2,4]OXADIAZOLIDIN-2-YL)-PROPIONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, Metabotropic glutamate receptor 5, ... | | Authors: | Koehl, A, Hu, H, Feng, D, Sun, B, Weis, W.I, Skiniotis, G.S, Mathiesen, J.M, Kobilka, B.K. | | Deposit date: | 2018-11-20 | | Release date: | 2019-01-23 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structural insights into the activation of metabotropic glutamate receptors.
Nature, 566, 2019
|
|
6N52
 
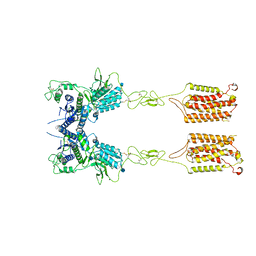 | | Metabotropic Glutamate Receptor 5 Apo Form | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Metabotropic glutamate receptor 5 | | Authors: | Koehl, A, Hu, H, Feng, D, Sun, B, Weis, W.I, Skiniotis, G.S, Mathiesen, J.M, Kobilka, B.K. | | Deposit date: | 2018-11-20 | | Release date: | 2019-01-23 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structural insights into the activation of metabotropic glutamate receptors.
Nature, 566, 2019
|
|
7VGE
 
 | | Structure of the PDZ deleted variant of HtrA2 protease (S306A) | | Descriptor: | Serine protease HTRA2, mitochondrial | | Authors: | Parui, A.L, Mishra, V, Bhaumik, P, Bose, K. | | Deposit date: | 2021-09-15 | | Release date: | 2022-06-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Inter-subunit crosstalk via PDZ synergistically governs allosteric activation of proapoptotic HtrA2.
Structure, 30, 2022
|
|
4GNK
 
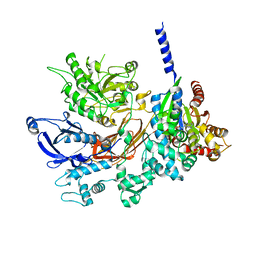 | | Crystal structure of Galphaq in complex with full-length human PLCbeta3 | | Descriptor: | 1-phosphatidylinositol 4,5-bisphosphate phosphodiesterase beta-3, CALCIUM ION, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Lyon, A.M, Tesmer, J.J.G. | | Deposit date: | 2012-08-17 | | Release date: | 2013-02-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Full-length G alpha (q)-phospholipase C-beta 3 structure reveals interfaces of the C-terminal coiled-coil domain.
Nat.Struct.Mol.Biol., 20, 2013
|
|
2ABJ
 
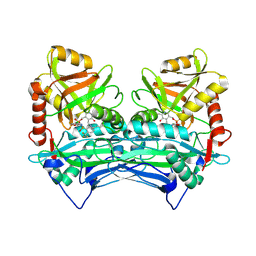 | | Crystal structure of human branched chain amino acid transaminase in a complex with an inhibitor, C16H10N2O4F3SCl, and pyridoxal 5' phosphate. | | Descriptor: | Branched-chain-amino-acid aminotransferase, cytosolic, N'-(5-CHLOROBENZOFURAN-2-CARBONYL)-2-(TRIFLUOROMETHYL)BENZENESULFONOHYDRAZIDE, ... | | Authors: | Ohren, J.F, Moreland, D.W, Rubin, J.R, Hu, H.L, McConnell, P.C, Mistry, A, Mueller, W.T, Scholten, J.D, Hasemann, C.H. | | Deposit date: | 2005-07-15 | | Release date: | 2006-06-27 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The design and synthesis of human branched-chain amino acid aminotransferase inhibitors for treatment of neurodegenerative diseases.
Bioorg.Med.Chem.Lett., 16, 2006
|
|
5EVM
 
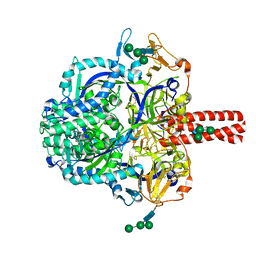 | | Crystal Structure of Nipah Virus Fusion Glycoprotein in the Prefusion State | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Fusion glycoprotein F0, MALONATE ION, ... | | Authors: | Xu, K, Nikolov, D.B. | | Deposit date: | 2015-11-20 | | Release date: | 2015-12-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.367 Å) | | Cite: | Crystal Structure of the Pre-fusion Nipah Virus Fusion Glycoprotein Reveals a Novel Hexamer-of-Trimers Assembly.
Plos Pathog., 11, 2015
|
|
4O6D
 
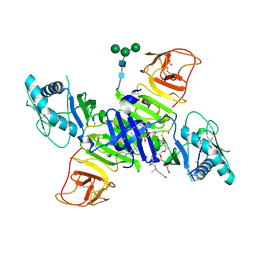 | | West Nile Virus Non-structural protein 1 (NS1) Form 1 crystal | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, NS1, ... | | Authors: | Akey, D.L, Smith, J.L. | | Deposit date: | 2013-12-20 | | Release date: | 2014-02-19 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.5936 Å) | | Cite: | Flavivirus NS1 structures reveal surfaces for associations with membranes and the immune system.
Science, 343, 2014
|
|
4O6B
 
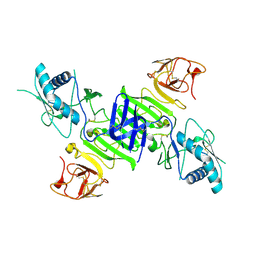 | |
4O6C
 
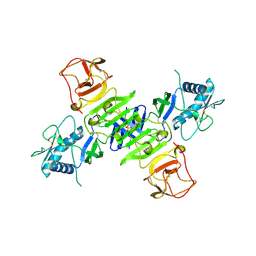 | |
2MCA
 
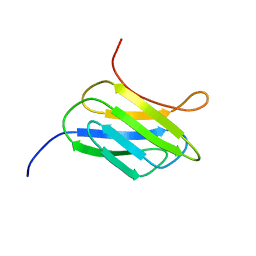 | | NMR structure of the protein YP_002937094.1 from Eubacterium rectale | | Descriptor: | Uncharacterized protein | | Authors: | Proudfoot, A, Serrano, P, Geralt, M, Dutta, S, Wuthrich, K, Joint Center for Structural Genomics (JCSG) | | Deposit date: | 2013-08-19 | | Release date: | 2013-09-04 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the protein YP_002937094.1 from Eubacterium rectale
To be Published
|
|
2MCT
 
 | | NMR structure of the protein ZP_02042476.1 from Ruminococcus gnavus | | Descriptor: | Uncharacterized protein | | Authors: | Martin, B.T, Serrano, P, Geralt, M, Dutta, S, Wuthrich, K, Joint Center for Structural Genomics (JCSG) | | Deposit date: | 2013-08-27 | | Release date: | 2013-11-13 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the protein ZP_02042476.1 from Ruminococcus gnavus.
To be Published
|
|
8AV8
 
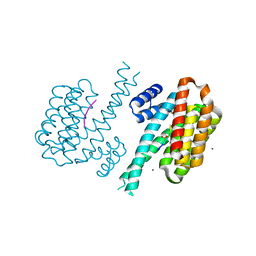 | | Small molecular stabilizer for ERalpha and 14-3-3 (1075300) | | Descriptor: | 14-3-3 protein sigma, 2-chloranyl-~{N}-[[1-(1-phenoxycyclopentyl)carbonylpiperidin-4-yl]methyl]ethanamide, Estrogen receptor, ... | | Authors: | Visser, E.J, Vandenboorn, E.M.F, Ottmann, C. | | Deposit date: | 2022-08-26 | | Release date: | 2023-09-06 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-Based Optimization of Covalent, Small-Molecule Stabilizers of the 14-3-3 sigma /ER alpha Protein-Protein Interaction from Nonselective Fragments.
J.Am.Chem.Soc., 145, 2023
|
|
8ALR
 
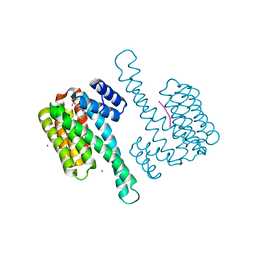 | | Small molecular stabilizer for ERalpha and 14-3-3 (1080272) | | Descriptor: | 14-3-3 protein sigma, 2-chloranyl-N-[[1-(1-phenylazanylcyclohexyl)carbonylpiperidin-4-yl]methyl]ethanamide, Estrogen receptor, ... | | Authors: | Visser, E.J, Vandenboorn, E.M.F, Ottmann, C. | | Deposit date: | 2022-08-01 | | Release date: | 2023-09-20 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure-Based Optimization of Covalent, Small-Molecule Stabilizers of the 14-3-3 sigma /ER alpha Protein-Protein Interaction from Nonselective Fragments.
J.Am.Chem.Soc., 145, 2023
|
|
8AQE
 
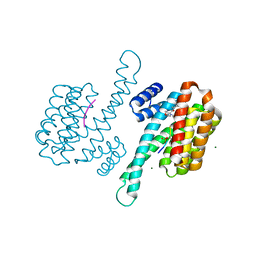 | | Small molecular stabilizer for ERalpha and 14-3-3 (1080295) | | Descriptor: | 14-3-3 protein sigma, 2-chloranyl-N-[7-[4-[(4-chlorophenyl)amino]oxan-4-yl]carbonyl-7-azaspiro[3.5]nonan-2-yl]ethanamide, Estrogen receptor, ... | | Authors: | Visser, E.J, Vandenboorn, E.M.F, Ottmann, C. | | Deposit date: | 2022-08-12 | | Release date: | 2023-09-20 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure-Based Optimization of Covalent, Small-Molecule Stabilizers of the 14-3-3 sigma /ER alpha Protein-Protein Interaction from Nonselective Fragments.
J.Am.Chem.Soc., 145, 2023
|
|
8AR4
 
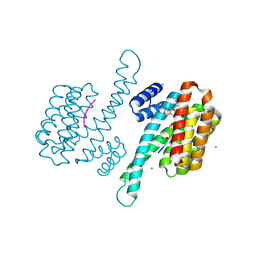 | | Small molecular stabilizer for ERalpha and 14-3-3 (1080300) | | Descriptor: | 14-3-3 protein sigma, 2-chloranyl-N-[[1-[4-[(4-chlorophenyl)amino]-2,2,6,6-tetramethyl-oxan-4-yl]carbonylpiperidin-4-yl]methyl]ethanamide, Estrogen receptor, ... | | Authors: | Visser, E.J, Vandenboorn, E.M.F, Ottmann, C. | | Deposit date: | 2022-08-15 | | Release date: | 2023-09-20 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure-Based Optimization of Covalent, Small-Molecule Stabilizers of the 14-3-3 sigma /ER alpha Protein-Protein Interaction from Nonselective Fragments.
J.Am.Chem.Soc., 145, 2023
|
|
8AI0
 
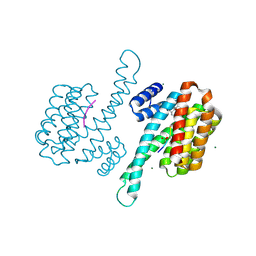 | | Small molecular stabilizer for ERalpha and 14-3-3sigma (1080268) | | Descriptor: | 14-3-3 protein sigma, 2-chloranyl-N-[[1-(2-methyl-2-phenylazanyl-propanoyl)piperidin-4-yl]methyl]ethanamide, Estrogen receptor, ... | | Authors: | Visser, E.J, Vandenboorn, E.M.F, Ottmann, C. | | Deposit date: | 2022-07-25 | | Release date: | 2023-09-20 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure-Based Optimization of Covalent, Small-Molecule Stabilizers of the 14-3-3 sigma /ER alpha Protein-Protein Interaction from Nonselective Fragments.
J.Am.Chem.Soc., 145, 2023
|
|
8ARX
 
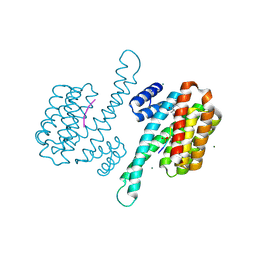 | | Small molecular stabilizer for ERalpha and 14-3-3sigma (1074378) | | Descriptor: | 14-3-3 protein sigma, 2-chloranyl-N-[[1-[1-(4-chloranylphenoxy)cyclopropyl]carbonylpiperidin-4-yl]methyl]ethanamide, Estrogen receptor, ... | | Authors: | Visser, E.J, Vandenboorn, E.M.F, Ottmann, C. | | Deposit date: | 2022-08-17 | | Release date: | 2023-09-20 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure-Based Optimization of Covalent, Small-Molecule Stabilizers of the 14-3-3 sigma /ER alpha Protein-Protein Interaction from Nonselective Fragments.
J.Am.Chem.Soc., 145, 2023
|
|
8AS1
 
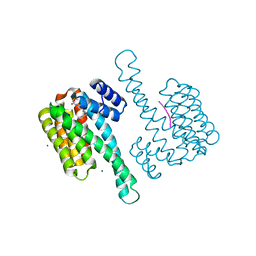 | | Small molecular stabilizer for ERalpha and 14-3-3 (1076398) | | Descriptor: | 14-3-3 protein sigma, 2-chloranyl-N-[[1-(4-phenylazanylpiperidin-4-yl)carbonylpiperidin-4-yl]methyl]ethanamide, Estrogen receptor, ... | | Authors: | Visser, E.J, Vandenboorn, E.M.F, Ottmann, C. | | Deposit date: | 2022-08-17 | | Release date: | 2023-09-20 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure-Based Optimization of Covalent, Small-Molecule Stabilizers of the 14-3-3 sigma /ER alpha Protein-Protein Interaction from Nonselective Fragments.
J.Am.Chem.Soc., 145, 2023
|
|
8ALV
 
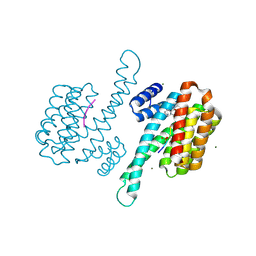 | | Small molecular stabilizer for ERalpha and 14-3-3 (1076403) | | Descriptor: | 14-3-3 protein sigma, 2-chloranyl-N-[[1-[1-[(4-chlorophenyl)amino]cyclohexyl]carbonylpiperidin-4-yl]methyl]ethanamide, Estrogen receptor, ... | | Authors: | Visser, E.J, Vandenboorn, E.M.F, Ottmann, C. | | Deposit date: | 2022-08-01 | | Release date: | 2023-09-20 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure-Based Optimization of Covalent, Small-Molecule Stabilizers of the 14-3-3 sigma /ER alpha Protein-Protein Interaction from Nonselective Fragments.
J.Am.Chem.Soc., 145, 2023
|
|
8AT9
 
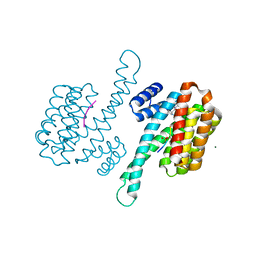 | | Small molecule stabilizer for ERalpha and 14-3-3 (1080269) | | Descriptor: | 14-3-3 protein sigma, 2-chloranyl-~{N}-[[1-(1-phenylazanylcyclobutyl)carbonylpiperidin-4-yl]methyl]ethanamide, Estrogen receptor, ... | | Authors: | Visser, E.J, Vandenboorn, E.M.F, Ottmann, C. | | Deposit date: | 2022-08-22 | | Release date: | 2023-09-20 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure-Based Optimization of Covalent, Small-Molecule Stabilizers of the 14-3-3 sigma /ER alpha Protein-Protein Interaction from Nonselective Fragments.
J.Am.Chem.Soc., 145, 2023
|
|
8AM7
 
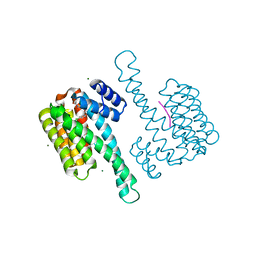 | | Small molecular stabilizer for ERalpha and 14-3-3 (1076397) | | Descriptor: | 14-3-3 protein sigma, 2-chloranyl-~{N}-[[1-[4-(4-chloranylphenoxy)piperidin-4-yl]carbonylpiperidin-4-yl]methyl]ethanamide, Estrogen receptor, ... | | Authors: | Visser, E.J, Vandenboorn, E.M.F, Ottmann, C. | | Deposit date: | 2022-08-02 | | Release date: | 2023-09-20 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure-Based Optimization of Covalent, Small-Molecule Stabilizers of the 14-3-3 sigma /ER alpha Protein-Protein Interaction from Nonselective Fragments.
J.Am.Chem.Soc., 145, 2023
|
|
8AQZ
 
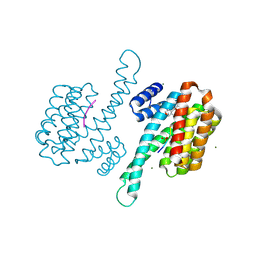 | | Small molecular stabilizer for ERalpha and 14-3-3 (1080267) | | Descriptor: | 14-3-3 protein sigma, 2-chloranyl-N-[2-[1-[4-[(4-chlorophenyl)amino]oxan-4-yl]carbonylpiperidin-4-yl]ethyl]ethanamide, Estrogen receptor, ... | | Authors: | Visser, E.J, Vandenboorn, E.M.F, Ottmann, C. | | Deposit date: | 2022-08-15 | | Release date: | 2023-09-20 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure-Based Optimization of Covalent, Small-Molecule Stabilizers of the 14-3-3 sigma /ER alpha Protein-Protein Interaction from Nonselective Fragments.
J.Am.Chem.Soc., 145, 2023
|
|
8ALW
 
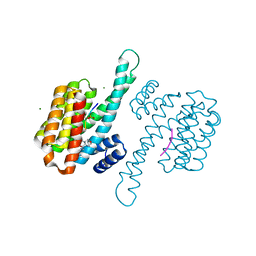 | | Small molecular stabilizer for ERalpha and 14-3-3 (1075310) | | Descriptor: | 14-3-3 protein sigma, 2-chloranyl-N-[[1-[4-[(4-chlorophenyl)amino]oxan-4-yl]carbonylpiperidin-4-yl]methyl]ethanamide, Estrogen receptor, ... | | Authors: | Visser, E.J, Vandenboorn, E.M.F, Ottmann, C. | | Deposit date: | 2022-08-01 | | Release date: | 2023-09-20 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure-Based Optimization of Covalent, Small-Molecule Stabilizers of the 14-3-3 sigma /ER alpha Protein-Protein Interaction from Nonselective Fragments.
J.Am.Chem.Soc., 145, 2023
|
|
