3KF9
 
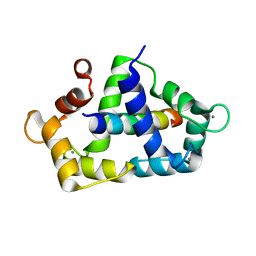 | | Crystal structure of the SdCen/skMLCK complex | | Descriptor: | CALCIUM ION, Caltractin, Myosin light chain kinase 2, ... | | Authors: | Radu, L, Assairi, L, Blouquit, Y, Durand, D, Miron, S, Charbonnier, J.B, Craescu, C.T. | | Deposit date: | 2009-10-27 | | Release date: | 2011-01-26 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural features of the complexes formed by Scherffelia dubia centrin
To be Published
|
|
2ZBK
 
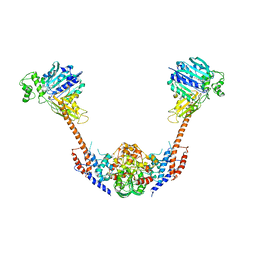 | | Crystal structure of an intact type II DNA topoisomerase: insights into DNA transfer mechanisms | | Descriptor: | RADICICOL, Type 2 DNA topoisomerase 6 subunit B, Type II DNA topoisomerase VI subunit A | | Authors: | Graille, M, Cladiere, L, Durand, D, Lecointe, F, Forterre, P, van Tilbeurgh, H, Paris-Sud Yeast Structural Genomics (YSG) | | Deposit date: | 2007-10-22 | | Release date: | 2008-02-12 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.56 Å) | | Cite: | Crystal Structure of an Intact Type II DNA Topoisomerase: Insights into DNA Transfer Mechanisms
Structure, 16, 2008
|
|
4ML3
 
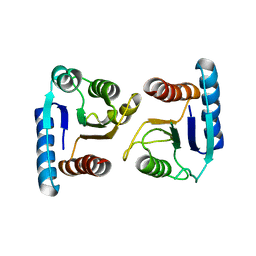 | | X-ray structure of ComE D58A REC domain from Streptococcus pneumoniae | | Descriptor: | Response regulator | | Authors: | Boudes, M, Sanchez, D, Durand, D, Graille, M, van Tilbeurgh, H, Quevillon-Cheruel, S. | | Deposit date: | 2013-09-06 | | Release date: | 2014-02-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Structural insights into the dimerization of the response regulator ComE from Streptococcus pneumoniae.
Nucleic Acids Res., 42, 2014
|
|
4MLD
 
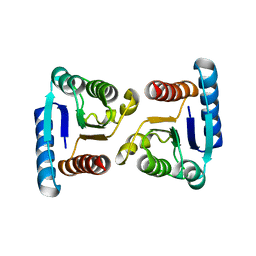 | | X-ray structure of ComE D58E REC domain from Streptococcus pneumoniae | | Descriptor: | Response regulator | | Authors: | Boudes, M, Sanchez, D, Durand, D, Graille, M, van Tilbeurgh, H, Quevillon-Cheruel, S. | | Deposit date: | 2013-09-06 | | Release date: | 2014-02-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.88 Å) | | Cite: | Structural insights into the dimerization of the response regulator ComE from Streptococcus pneumoniae.
Nucleic Acids Res., 42, 2014
|
|
6GW7
 
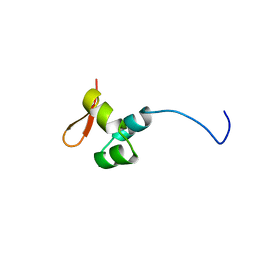 | | The CTD of HpDprA, a DNA binding Winged Helix domain which do not bind dsDNA | | Descriptor: | DNA protecting protein DprA | | Authors: | Lisboa, J, Celma, L, Sanchez, D, Marquis, M, Andreani, J, Guerois, R, Ochsenbein, F, Durand, D, Marsin, S, Cuniasse, P, Radicella, J.P, Quevillon-Cheruel, S. | | Deposit date: | 2018-06-22 | | Release date: | 2019-04-24 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | The C-terminal domain of HpDprA is a DNA-binding winged helix domain that does not bind double-stranded DNA.
Febs J., 286, 2019
|
|
2VDU
 
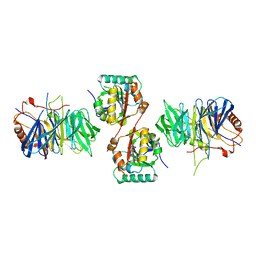 | | Structure of trm8-trm82, THE YEAST TRNA m7G methylation complex | | Descriptor: | PHOSPHATE ION, TRNA (GUANINE-N(7)-)-METHYLTRANSFERASE, TRNA (GUANINE-N(7)-)-METHYLTRANSFERASE-ASSOCIATED WD REPEAT PROTEIN TRM82 | | Authors: | Leulliot, N, Chaillet, M, Durand, D, Ulryck, N, Blondeau, K, Van Tilbeurgh, H. | | Deposit date: | 2007-10-11 | | Release date: | 2007-12-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of the Yeast tRNA M7G Methylation Complex.
Structure, 16, 2008
|
|
2W9M
 
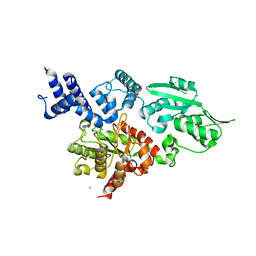 | | Structure of family X DNA polymerase from Deinococcus radiodurans | | Descriptor: | MERCURY (II) ION, POLYMERASE X, ZINC ION | | Authors: | Leulliot, N, Cladiere, L, Lecointe, F, Durand, D, Hubscher, U, van Tilbeurgh, H. | | Deposit date: | 2009-01-27 | | Release date: | 2009-02-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | The Family X DNA Polymerase from Deinococcus Radioduran Adopts a Non-Standard Extended Conformation.
J.Biol.Chem., 284, 2009
|
|
2VDV
 
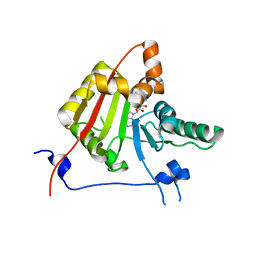 | | Structure of trm8, m7G methylation enzyme | | Descriptor: | S-ADENOSYLMETHIONINE, TRNA (GUANINE-N(7)-)-METHYLTRANSFERASE | | Authors: | Leulliot, N, Chaillet, M, Durand, D, Ulryck, N, Blondeau, K, Van Tilbeurgh, H. | | Deposit date: | 2007-10-11 | | Release date: | 2007-12-18 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of the Yeast tRNA M7G Methylation Complex.
Structure, 16, 2008
|
|
4AKK
 
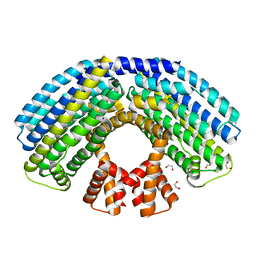 | | Structure of the NasR transcription antiterminator | | Descriptor: | 1,2-ETHANEDIOL, NITRATE REGULATORY PROTEIN | | Authors: | Boudes, M, Lazar, N, Graille, M, Durand, D, Gaidenko, T.A, Stewart, V, van Tilbeurgh, H. | | Deposit date: | 2012-02-24 | | Release date: | 2012-06-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.145 Å) | | Cite: | The Structure of the Nasr Transcription Antiterminator Reveals a One-Component System with a Nit Nitrate Receptor Coupled to an Antar RNA-Binding Effector.
Mol.Microbiol., 85, 2012
|
|
4AK8
 
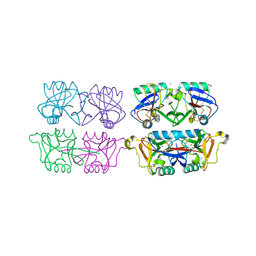 | | Structure of F241L mutant of langerin carbohydrate recognition domain. | | Descriptor: | C-TYPE LECTIN DOMAIN FAMILY 4 MEMBER K, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Chabrol, E, Thepaut, M, Dezutter-Dambuyant, C, Vives, C, Marcoux, J, Kahn, R, Valadeau-Guilemond, J, Vachette, P, Durand, D, Fieschi, F. | | Deposit date: | 2012-02-22 | | Release date: | 2013-04-03 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Alteration of the Langerin Oligomerization State Affects Birbeck Granule Formation.
Biophys.J., 108, 2015
|
|
6GWJ
 
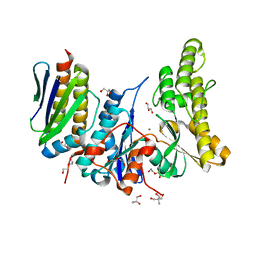 | | Human OSGEP / LAGE3 / GON7 complex | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DI(HYDROXYETHYL)ETHER, EKC/KEOPS complex subunit GON7, ... | | Authors: | Missoury, S, Liger, D, Durand, D, Collinet, B, Tilbeurgh, V.H. | | Deposit date: | 2018-06-25 | | Release date: | 2019-07-03 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Defects in t 6 A tRNA modification due to GON7 and YRDC mutations lead to Galloway-Mowat syndrome.
Nat Commun, 10, 2019
|
|
4A8E
 
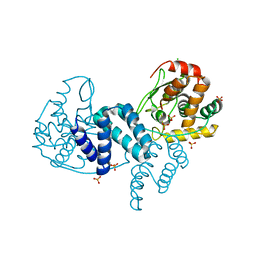 | | The structure of a dimeric Xer recombinase from archaea | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, PROBABLE TYROSINE RECOMBINASE XERC-LIKE, ... | | Authors: | Brooks, M.A, ElArnaout, T, Duranda, D, Lisboa, J, Lazar, N, Raynal, B, vanTilbeurgh, H, Serre, M, Quevillon-Cheruel, S. | | Deposit date: | 2011-11-21 | | Release date: | 2012-12-05 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | The Carboxy-Terminal Alpha N Helix of the Archaeal Xera Tyrosine Recombinase is a Molecular Switch to Control Site-Specific Recombination.
Plos One, 8, 2013
|
|
3P26
 
 | | Crystal structure of S. cerevisiae Hbs1 protein (apo-form), a translational GTPase involved in RNA quality control pathways and interacting with Dom34/Pelota | | Descriptor: | Elongation factor 1 alpha-like protein | | Authors: | van den Elzen, A, Henri, J, Lazar, N, Gas, M.E, Durand, D, Lacroute, F, Nicaise, M, van Tilbeurgh, H, Sraphin, B, Graille, M, Paris-Sud Yeast Structural Genomics (YSG) | | Deposit date: | 2010-10-01 | | Release date: | 2010-11-17 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Dissection of Dom34-Hbs1 reveals independent functions in two RNA quality control pathways.
Nat.Struct.Mol.Biol., 17, 2010
|
|
3P27
 
 | | Crystal structure of S. cerevisiae Hbs1 protein (GDP-bound form), a translational GTPase involved in RNA quality control pathways and interacting with Dom34/Pelota | | Descriptor: | Elongation factor 1 alpha-like protein, GUANOSINE-5'-DIPHOSPHATE | | Authors: | van den Elzen, A, Henri, J, Lazar, N, Gas, M.E, Durand, D, Lacroute, F, Nicaise, M, van Tilbeurgh, H, Sraphin, B, Graille, M, Paris-Sud Yeast Structural Genomics (YSG) | | Deposit date: | 2010-10-01 | | Release date: | 2010-11-17 | | Last modified: | 2012-03-14 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Dissection of Dom34-Hbs1 reveals independent functions in two RNA quality control pathways.
Nat.Struct.Mol.Biol., 17, 2010
|
|
4CBV
 
 | | X-ray structure of full-length ComE from Streptococcus pneumoniae. | | Descriptor: | COME | | Authors: | Boudes, M, Durand, D, Graille, M, van Tilbeurgh, H, Quevillon-Cheruel, S. | | Deposit date: | 2013-10-16 | | Release date: | 2014-02-12 | | Last modified: | 2014-05-14 | | Method: | X-RAY DIFFRACTION (3.39 Å) | | Cite: | Structural Insights Into the Dimerization of the Response Regulator Come from Streptococcus Pneumoniae.
Nucleic Acids Res., 42, 2014
|
|
4YDU
 
 | | Crystal structure of E. coli YgjD-YeaZ heterodimer in complex with ADP | | Descriptor: | ACETATE ION, ADENOSINE-5'-DIPHOSPHATE, FE (III) ION, ... | | Authors: | Zhang, W, Collinet, B, Perrochia, L, Durand, D, Van Tilbeurgh, H. | | Deposit date: | 2015-02-23 | | Release date: | 2015-03-04 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | The ATP-mediated formation of the YgjD-YeaZ-YjeE complex is required for the biosynthesis of tRNA t6A in Escherichia coli.
Nucleic Acids Res., 43, 2015
|
|
3UQZ
 
 | |
6NAK
 
 | | BACTERIAL PROTEIN COMPLEX TM BDE complex | | Descriptor: | DIPHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER, MAGNESIUM ION, TsaE, ... | | Authors: | Stec, B. | | Deposit date: | 2018-12-05 | | Release date: | 2023-04-26 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.14 Å) | | Cite: | Discovery of the Universal tRNA Binding Mode for the TsaD-like Components of the t6A tRNA Modification Pathway
Biophysica, 3, 2023
|
|
4OJJ
 
 | | Structure of C-terminal domain from S. cerevisiae Pat1 decapping activator (Space group : P212121) | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DNA topoisomerase 2-associated protein PAT1, ... | | Authors: | Fourati-Kammoun, Z, Kolesnikova, O, Back, R, Keller, J, Lazar, N, Gaudon-Plesse, C, Seraphin, B, Graille, M. | | Deposit date: | 2014-01-21 | | Release date: | 2014-10-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | The C-terminal domain from S. cerevisiae Pat1 displays two conserved regions involved in decapping factor recruitment.
Plos One, 9, 2014
|
|
4OGP
 
 | | Structure of C-terminal domain from S. cerevisiae Pat1 decapping activator (Space group : P21) | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, DNA topoisomerase 2-associated protein PAT1 | | Authors: | Fourati-Kammoun, Z, Kolesnikova, O, Back, R, Keller, J, Lazar, N, Gaudon-Plesse, C, Seraphin, B, Graille, M. | | Deposit date: | 2014-01-16 | | Release date: | 2014-10-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The C-terminal domain from S. cerevisiae Pat1 displays two conserved regions involved in decapping factor recruitment.
Plos One, 9, 2014
|
|
4ZV6
 
 | | Crystal structure of the artificial alpharep-7 octarellinV.1 complex | | Descriptor: | AlphaRep-7, Octarellin V.1 | | Authors: | Figueroa, M, Sleutel, M, Urvoas, A, Valerio-Lepiniec, M, Minard, P, Martial, J.A, van de Weerdt, C. | | Deposit date: | 2015-05-18 | | Release date: | 2016-05-25 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | The unexpected structure of the designed protein Octarellin V.1 forms a challenge for protein structure prediction tools.
J.Struct.Biol., 195, 2016
|
|
5BOP
 
 | | Crystal structure of the artificial nanobody octarellinV.1 complex | | Descriptor: | Nanobody, Octarellin V.1 | | Authors: | Figueroa, M, Sleutel, M, Pardon, E, Steyaert, J, Martial, J.A, van de Weerdt, C. | | Deposit date: | 2015-05-27 | | Release date: | 2016-05-25 | | Last modified: | 2016-06-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The unexpected structure of the designed protein Octarellin V.1 forms a challenge for protein structure prediction tools.
J.Struct.Biol., 195, 2016
|
|
6YNU
 
 | | CaM-P458 complex (crystal form 1) | | Descriptor: | Bifunctional adenylate cyclase toxin/hemolysin CyaA, CALCIUM ION, Calmodulin-1 | | Authors: | Mechaly, A.E, Voegele, A, Haouz, A, Chenal, A. | | Deposit date: | 2020-04-14 | | Release date: | 2021-03-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.12 Å) | | Cite: | A High-Affinity Calmodulin-Binding Site in the CyaA Toxin Translocation Domain is Essential for Invasion of Eukaryotic Cells.
Adv Sci, 8, 2021
|
|
6YNS
 
 | | CaM-P458 complex (crystal form 2) | | Descriptor: | Bifunctional adenylate cyclase toxin/hemolysin CyaA, CALCIUM ION, Calmodulin-1 | | Authors: | Mechaly, A.E, Voegele, A, Haouz, A, Chenal, A. | | Deposit date: | 2020-04-14 | | Release date: | 2021-03-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.94 Å) | | Cite: | A High-Affinity Calmodulin-Binding Site in the CyaA Toxin Translocation Domain is Essential for Invasion of Eukaryotic Cells.
Adv Sci, 8, 2021
|
|
5TJT
 
 | |
