6Q1Q
 
 | |
6Q1S
 
 | |
6Q1R
 
 | |
5L1P
 
 | | X-ray Structure of Cytochrome P450 PntM with Pentalenolactone | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, Pentalenolactone synthase, pentalenolactone | | Authors: | Duan, L, Jogl, G, Cane, D.E. | | Deposit date: | 2016-07-29 | | Release date: | 2016-09-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | The Cytochrome P450-Catalyzed Oxidative Rearrangement in the Final Step of Pentalenolactone Biosynthesis: Substrate Structure Determines Mechanism.
J.Am.Chem.Soc., 138, 2016
|
|
5L1O
 
 | | X-ray Structure of Cytochrome P450 PntM with Pentalenolactone F | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, Pentalenolactone synthase, pentalenolactone F | | Authors: | Duan, L, Jogl, G, Cane, D.E. | | Deposit date: | 2016-07-29 | | Release date: | 2016-09-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | The Cytochrome P450-Catalyzed Oxidative Rearrangement in the Final Step of Pentalenolactone Biosynthesis: Substrate Structure Determines Mechanism.
J.Am.Chem.Soc., 138, 2016
|
|
5L1Q
 
 | | X-ray Structure of Cytochrome P450 PntM with Dihydropentalenolactone F | | Descriptor: | Dihydropentalenolactone F, PROTOPORPHYRIN IX CONTAINING FE, Pentalenolactone synthase | | Authors: | Duan, L, Jogl, G, Cane, D.E. | | Deposit date: | 2016-07-29 | | Release date: | 2016-09-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | The Cytochrome P450-Catalyzed Oxidative Rearrangement in the Final Step of Pentalenolactone Biosynthesis: Substrate Structure Determines Mechanism.
J.Am.Chem.Soc., 138, 2016
|
|
5L1S
 
 | |
5L1W
 
 | |
5L1V
 
 | |
5L1R
 
 | | X-ray Structure of the Substrate-free Cytochrome P450 PntM | | Descriptor: | BICINE, PROTOPORPHYRIN IX CONTAINING FE, Pentalenolactone synthase | | Authors: | Duan, L, Jogl, G, Cane, D.E. | | Deposit date: | 2016-07-29 | | Release date: | 2016-09-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Cytochrome P450-Catalyzed Oxidative Rearrangement in the Final Step of Pentalenolactone Biosynthesis: Substrate Structure Determines Mechanism.
J.Am.Chem.Soc., 138, 2016
|
|
5L1U
 
 | |
5L1T
 
 | |
5FID
 
 | |
5V93
 
 | | Cryo-EM structure of the 70S ribosome from Mycobacterium tuberculosis bound with Capreomycin | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Yang, K, Chang, J.-Y, Cui, Z, Li, X, Meng, R, Duan, L, Thongchol, J, Jakana, J, Huwe, C, Sacchettini, J, Zhang, J. | | Deposit date: | 2017-03-22 | | Release date: | 2017-09-20 | | Last modified: | 2020-08-12 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structural insights into species-specific features of the ribosome from the human pathogen Mycobacterium tuberculosis.
Nucleic Acids Res., 45, 2017
|
|
8K20
 
 | | Cryo-EM structure of KEOPS complex from Arabidopsis thaliana | | Descriptor: | At4g34412, At5g53043, FE (III) ION, ... | | Authors: | Zheng, X.X, Zhu, L, Duan, L, Zhang, W.H. | | Deposit date: | 2023-07-11 | | Release date: | 2024-04-03 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Molecular basis of A. thaliana KEOPS complex in biosynthesizing tRNA t6A.
Nucleic Acids Res., 52, 2024
|
|
2A6A
 
 | |
7SIU
 
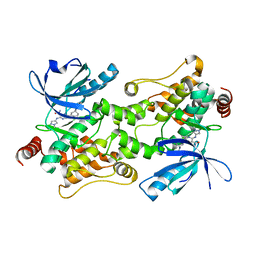 | | Crystal structure of HPK1 (MAP4K1) complex with inhibitor A-745 | | Descriptor: | 6-[(5R)-5-benzamidocyclohex-1-en-1-yl]-3-[(1-methyl-1H-pyrazol-4-yl)amino]pyrazine-2-carboxamide, Mitogen-activated protein kinase kinase kinase kinase 1, SULFATE ION | | Authors: | Longenecker, K.L, Korepanova, A, Qiu, W. | | Deposit date: | 2021-10-14 | | Release date: | 2022-03-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.786 Å) | | Cite: | The HPK1 Inhibitor A-745 Verifies the Potential of Modulating T Cell Kinase Signaling for Immunotherapy.
Acs Chem.Biol., 17, 2022
|
|
3L5O
 
 | |
3KK7
 
 | |
3NL9
 
 | |
2RA9
 
 | |
2QTP
 
 | |
2RE3
 
 | |
3B77
 
 | |
3BYQ
 
 | |
