7B6K
 
 | | Crystal structure of MurE from E.coli in complex with Z57715447 | | Descriptor: | 5-cyclohexyl-3-(pyridin-4-yl)-1,2,4-oxadiazole, CITRIC ACID, DIMETHYL SULFOXIDE, ... | | Authors: | Koekemoer, L, Steindel, M, Fairhead, M, Talon, R, Douangamath, A, Arrowsmith, C.H, Edwards, A.M, Bountra, C, von Delft, F, Krojer, T, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-12-07 | | Release date: | 2020-12-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.838 Å) | | Cite: | Crystal structure of MurE from E.coli
To Be Published
|
|
7B6I
 
 | | Crystal structure of MurE from E.coli in complex with Z1373445602 | | Descriptor: | 4-(3-fluoranylpyridin-2-yl)-1-methyl-piperazin-2-one, CITRIC ACID, ISOPROPYL ALCOHOL, ... | | Authors: | Koekemoer, L, Steindel, M, Fairhead, M, Talon, R, Douangamath, A, Arrowsmith, C.H, Edwards, A.M, Bountra, C, von Delft, F, Krojer, T, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-12-07 | | Release date: | 2020-12-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.069 Å) | | Cite: | Crystal structure of MurE from E.coli
To Be Published
|
|
6Q8Z
 
 | | Structure of human galactokinase 1 bound with N-(Cyclobutylmethyl)-1,5-dimethyl-1H-pyrazole-4-carboxamide | | Descriptor: | 2-(1,3-benzoxazol-2-ylamino)spiro[1,6,7,8-tetrahydroquinazoline-4,1'-cyclohexane]-5-one, Galactokinase, beta-D-galactopyranose, ... | | Authors: | Mackinnon, S.R, Bezerra, G.A, Zhang, M, Foster, W, Krojer, T, Brandao-Neto, J, Douangamath, A, Arrowsmith, C, Edwards, A, Bountra, C, Brennan, P, Lai, K, Yue, W.W. | | Deposit date: | 2018-12-16 | | Release date: | 2019-01-23 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of human galactokinase 1 bound with N-(Cyclobutylmethyl)-1,5-dimethyl-1H-pyrazole-4-carboxamide
To Be Published
|
|
6Q91
 
 | | Structure of human galactokinase 1 bound with 5-Chloro-N-isobutyl-2-methoxybenzamide | | Descriptor: | 2-(1,3-benzoxazol-2-ylamino)spiro[1,6,7,8-tetrahydroquinazoline-4,1'-cyclohexane]-5-one, 5-chloranyl-2-methoxy-~{N}-(2-methylpropyl)benzamide, Galactokinase, ... | | Authors: | Mackinnon, S.R, Bezerra, G.A, Zhang, M, Foster, W, Krojer, T, Brandao-Neto, J, Douangamath, A, Arrowsmith, C, Edwards, A, Bountra, C, Brennan, P, Lai, K, Yue, W.W. | | Deposit date: | 2018-12-17 | | Release date: | 2019-01-23 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of human galactokinase 1 bound with 5-Chloro-N-isobutyl-2-methoxybenzamide
To Be Published
|
|
6Q3W
 
 | | Structure of human galactokinase 1 bound with Ethyl 1-(2-pyrazinyl)-4-piperidinecarboxylate | | Descriptor: | 2-(1,3-benzoxazol-2-ylamino)spiro[1,6,7,8-tetrahydroquinazoline-4,1'-cyclohexane]-5-one, Galactokinase, beta-D-galactopyranose, ... | | Authors: | Mackinnon, S.R, Bezerra, G.A, Zhang, M, Foster, W, Krojer, T, Brandao-Neto, J, Douangamath, A, Arrowsmith, C, Edwards, A, Bountra, C, Brennan, P, Lai, K, Yue, W.W. | | Deposit date: | 2018-12-04 | | Release date: | 2019-01-23 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.962 Å) | | Cite: | Structure of human galactokinase 1 bound with Ethyl 1-(2-pyrazinyl)-4-piperidinecarboxylate
To Be Published
|
|
7R4N
 
 | | Structure of human hydroxyacid oxidase 1 bound with 5-bromo-N-methyl-1H-indazole-3-carboxamide | | Descriptor: | 1,2-ETHANEDIOL, 5-bromanyl-N-methyl-1H-indazole-3-carboxamide, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Mackinnon, S, Bezerra, G.A, Krojer, T, Bradley, A.R, Talon, R, Brandeo-Neto, J, Douangamath, A, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Oppermann, U, Brennan, P.E, Yue, W.W. | | Deposit date: | 2022-02-08 | | Release date: | 2022-03-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of human hydroxyacid oxidase 1 bound with 5-bromo-N-methyl-1H-indazole-3-carboxamide
To Be Published
|
|
7R4O
 
 | | Structure of human hydroxyacid oxidase 1 bound with 2-((4H-1,2,4-triazol-3-yl)thio)-1-(4-(3-chlorophenyl)piperazin-1-yl)ethan-1-one | | Descriptor: | 2-((4H-1,2,4-triazol-3-yl)thio)-1-(4-(3-chlorophenyl)piperazin-1-yl)ethan-1-one, FLAVIN MONONUCLEOTIDE, Hydroxyacid oxidase 1 | | Authors: | Mackinnon, S, Bezerra, G.A, Krojer, T, Bradley, A.R, Talon, R, Brandeo-Neto, J, Douangamath, A, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Oppermann, U, Brennan, P.E, Yue, W.W. | | Deposit date: | 2022-02-08 | | Release date: | 2022-05-04 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure of human hydroxyacid oxidase 1 bound with 5-bromo-N-methyl-1H-indazole-3-carboxamide
To Be Published
|
|
7R4P
 
 | | Structure of human hydroxyacid oxidase 1 bound with 6-amino-1-benzyl-5-(methylamino)pyrimidine-2,4(1H,3H)-dione | | Descriptor: | 6-amino-1-benzyl-5-(methylamino)pyrimidine-2,4(1H,3H)-dione, FLAVIN MONONUCLEOTIDE, Hydroxyacid oxidase 1 | | Authors: | Mackinnon, S, Bezerra, G.A, Krojer, T, Bradley, A.R, Talon, R, Brandeo-Neto, J, Douangamath, A, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Oppermann, U, Brennan, P.E, Yue, W.W. | | Deposit date: | 2022-02-09 | | Release date: | 2022-05-04 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Structure of human hydroxyacid oxidase 1 bound with 5-bromo-N-methyl-1H-indazole-3-carboxamide
To Be Published
|
|
6QJE
 
 | | Structure of human galactokinase 1 bound with 4-{[2-(Methylsulfonyl)-1H-imidazol-1-yl]methyl}-1,3-thiazole | | Descriptor: | 2-(1,3-benzoxazol-2-ylamino)spiro[1,6,7,8-tetrahydroquinazoline-4,1'-cyclohexane]-5-one, 4-[(2-methylsulfonylimidazol-1-yl)methyl]-1,3-thiazole, Galactokinase, ... | | Authors: | Mackinnon, S.R, Bezerra, G.A, Zhang, M, Foster, W, Krojer, T, Brandao-Neto, J, Douangamath, A, Arrowsmith, C, Edwards, A, Bountra, C, Brennan, P, Lai, K, Yue, W.W. | | Deposit date: | 2019-01-24 | | Release date: | 2019-02-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of human galactokinase 1 bound with 4-{[2-(Methylsulfonyl)-1H-imidazol-1-yl]methyl}-1,3-thiazole
To Be Published
|
|
6Q90
 
 | | Structure of human galactokinase 1 bound with 1-(4-Methoxyphenyl)-3-(4-pyridinyl)urea | | Descriptor: | 2-(1,3-benzoxazol-2-ylamino)spiro[1,6,7,8-tetrahydroquinazoline-4,1'-cyclohexane]-5-one, Galactokinase, N-(4-methoxyphenyl)-N'-pyridin-4-ylurea, ... | | Authors: | Mackinnon, S.R, Bezerra, G.A, Zhang, M, Foster, W, Krojer, T, Brandao-Neto, J, Douangamath, A, Arrowsmith, C, Edwards, A, Bountra, C, Brennan, P, Lai, K, Yue, W.W. | | Deposit date: | 2018-12-16 | | Release date: | 2019-01-23 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of human galactokinase 1 bound with 1-(4-Methoxyphenyl)-3-(4-pyridinyl)urea
To Be Published
|
|
7B9E
 
 | | Crystal structure of MurE from E.coli in complex with Z275151340 | | Descriptor: | 4-chloro-N-cyclopentyl-1-methyl-1H-pyrazole-3-carboxamide, ISOPROPYL ALCOHOL, UDP-N-acetylmuramoyl-L-alanyl-D-glutamate-2,6-diaminopimelate ligase | | Authors: | Koekemoer, L, Steindel, M, Fairhead, M, Talon, R, Douangamath, A, Arrowsmith, C.H, Edwards, A.M, Bountra, C, von Delft, F, Krojer, T, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-12-14 | | Release date: | 2020-12-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Crystal structure of MurE from E.coli
To Be Published
|
|
7AEG
 
 | | SARS-CoV-2 main protease in a covalent complex with SDZ 224015 derivative, compound 5 | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE, N-[(benzyloxy)carbonyl]-L-valyl-N-[(1S)-1-(carboxymethyl)-3-fluoro-2-oxopropyl]-L-alaninamide | | Authors: | Owen, C.D, Redhead, M.A, Lukacik, P, Strain-Damerell, C, Fearon, D, Brewitz, L, Collette, A, Robinson, C, Collins, P, Radoux, C, Navratilova, I, Douangamath, A, von Delft, F, Malla, T.R, Nugen, T, Hull, H, Tumber, A, Schofield, C.J, Hallet, D, Stuart, D.I, Hopkins, A.L, Walsh, M.A. | | Deposit date: | 2020-09-17 | | Release date: | 2021-07-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Bispecific repurposed medicines targeting the viral and immunological arms of COVID-19.
Sci Rep, 11, 2021
|
|
6ZSL
 
 | | Crystal structure of the SARS-CoV-2 helicase at 1.94 Angstrom resolution | | Descriptor: | PHOSPHATE ION, SARS-CoV-2 helicase NSP13, ZINC ION | | Authors: | Newman, J.A, Yosaatmadja, Y, Douangamath, A, Arrowsmith, C.H, von Delft, F, Edwards, A, Bountra, C, Gileadi, O. | | Deposit date: | 2020-07-15 | | Release date: | 2020-07-29 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Structure, mechanism and crystallographic fragment screening of the SARS-CoV-2 NSP13 helicase.
Nat Commun, 12, 2021
|
|
6Y84
 
 | | SARS-CoV-2 main protease with unliganded active site (2019-nCoV, coronavirus disease 2019, COVID-19) | | Descriptor: | 3C-like proteinase nsp5, DIMETHYL SULFOXIDE | | Authors: | Owen, C.D, Lukacik, P, Strain-Damerell, C.M, Douangamath, A, Powell, A.J, Fearon, D, Brandao-Neto, J, Crawshaw, A.D, Aragao, D, Williams, M, Flaig, R, Hall, D.R, McAuley, K.E, Mazzorana, M, Stuart, D.I, von Delft, F, Walsh, M.A. | | Deposit date: | 2020-03-03 | | Release date: | 2020-03-11 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | COVID-19 main protease with unliganded active site
To Be Published
|
|
7N7W
 
 | | Crystal Structure of SARS-CoV-2 NendoU in complex with CSC000178569 | | Descriptor: | N-(2-fluorophenyl)-N'-methylurea, Uridylate-specific endoribonuclease | | Authors: | Godoy, A.S, Douangamath, A, Nakamura, A.M, Dias, A, Krojer, T, Noske, G.D, Gawiljuk, V.O, Fernandes, R.S, Fairhead, M, Powell, A, Dunnet, L, Aimon, A, Fearon, D, Brandao-Neto, J, Skyner, R, von Delft, F, Oliva, G. | | Deposit date: | 2021-06-11 | | Release date: | 2021-09-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Allosteric regulation and crystallographic fragment screening of SARS-CoV-2 NSP15 endoribonuclease.
Nucleic Acids Res., 51, 2023
|
|
7N83
 
 | | Crystal Structure of SARS-CoV-2 NendoU in complex with Z2443429438 | | Descriptor: | (3S)-1-(phenylsulfonyl)pyrrolidin-3-amine, CITRIC ACID, Uridylate-specific endoribonuclease | | Authors: | Godoy, A.S, Douangamath, A, Nakamura, A.M, Dias, A, Krojer, T, Noske, G.D, Gawiljuk, V.O, Fernandes, R.S, Fairhead, M, Powell, A, Dunnet, L, Aimon, A, Fearon, D, Brandao-Neto, J, Skyner, R, von Delft, F, Oliva, G. | | Deposit date: | 2021-06-12 | | Release date: | 2021-09-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Allosteric regulation and crystallographic fragment screening of SARS-CoV-2 NSP15 endoribonuclease.
Nucleic Acids Res., 51, 2023
|
|
7N7U
 
 | | Crystal Structure of SARS-CoV-2 NendoU in complex with LIZA-7 | | Descriptor: | 1-[(2~{R},4~{S},5~{R})-5-[[(azanylidene-$l^{4}-azanylidene)amino]methyl]-4-oxidanyl-oxolan-2-yl]-5-methyl-pyrimidine-2,4-dione, Uridylate-specific endoribonuclease | | Authors: | Godoy, A.S, Douangamath, A, Nakamura, A.M, Dias, A, Krojer, T, Noske, G.D, Gawiljuk, V.O, Fernandes, R.S, Fairhead, M, Powell, A, Dunnet, L, Aimon, A, Fearon, D, Brandao-Neto, J, Skyner, R, von Delft, F, Oliva, G. | | Deposit date: | 2021-06-11 | | Release date: | 2021-09-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Allosteric regulation and crystallographic fragment screening of SARS-CoV-2 NSP15 endoribonuclease.
Nucleic Acids Res., 51, 2023
|
|
7N7Y
 
 | | Crystal Structure of SARS-CoV-2 NendoU in complex with Z18197050 | | Descriptor: | Uridylate-specific endoribonuclease, methyl 4-sulfamoylbenzoate | | Authors: | Godoy, A.S, Douangamath, A, Nakamura, A.M, Dias, A, Krojer, T, Noske, G.D, Gawiljuk, V.O, Fernandes, R.S, Fairhead, M, Powell, A, Dunnet, L, Aimon, A, Fearon, D, Brandao-Neto, J, Skyner, R, von Delft, F, Oliva, G. | | Deposit date: | 2021-06-11 | | Release date: | 2021-09-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Allosteric regulation and crystallographic fragment screening of SARS-CoV-2 NSP15 endoribonuclease.
Nucleic Acids Res., 51, 2023
|
|
7N7R
 
 | | Crystal Structure of SARS-CoV-2 NendoU in complex with Z2472938267 | | Descriptor: | 1-[2-(2-oxidanylidenepyrrolidin-1-yl)ethyl]-3-phenyl-urea, Uridylate-specific endoribonuclease | | Authors: | Godoy, A.S, Douangamath, A, Nakamura, A.M, Dias, A, Krojer, T, Noske, G.D, Gawiljuk, V.O, Fernandes, R.S, Fairhead, M, Powell, A, Dunnet, L, Aimon, A, Fearon, D, Brandao-Neto, J, Skyner, R, von Delft, F, Oliva, G. | | Deposit date: | 2021-06-11 | | Release date: | 2021-09-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Allosteric regulation and crystallographic fragment screening of SARS-CoV-2 NSP15 endoribonuclease.
Nucleic Acids Res., 51, 2023
|
|
7NIO
 
 | | Crystal structure of the SARS-CoV-2 helicase APO form | | Descriptor: | SARS-CoV-2 helicase NSP13, ZINC ION | | Authors: | Newman, J.A, Yosaatmadja, Y, Douangamath, A, Bountra, C, Gileadi, O. | | Deposit date: | 2021-02-12 | | Release date: | 2021-03-17 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure, mechanism and crystallographic fragment screening of the SARS-CoV-2 NSP13 helicase.
Nat Commun, 12, 2021
|
|
7NN0
 
 | | Crystal structure of the SARS-CoV-2 helicase in complex with AMP-PNP | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, SARS-CoV-2 helicase NSP13, ... | | Authors: | Newman, J.A, Yosaatmadja, Y, Douangamath, A, Bountra, C, Gileadi, O. | | Deposit date: | 2021-02-23 | | Release date: | 2021-03-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.04 Å) | | Cite: | Structure, mechanism and crystallographic fragment screening of the SARS-CoV-2 NSP13 helicase.
Nat Commun, 12, 2021
|
|
7NNG
 
 | | Crystal structure of the SARS-CoV-2 helicase in complex with Z2327226104 | | Descriptor: | 1-(2-methylphenyl)-1,2,3-triazole-4-carboxylic acid, PHOSPHATE ION, SARS-CoV-2 helicase NSP13, ... | | Authors: | Newman, J.A, Yosaatmadja, Y, Douangamath, A, Bountra, C, Gileadi, O. | | Deposit date: | 2021-02-24 | | Release date: | 2021-04-07 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Structure, mechanism and crystallographic fragment screening of the SARS-CoV-2 NSP13 helicase.
Nat Commun, 12, 2021
|
|
7MBG
 
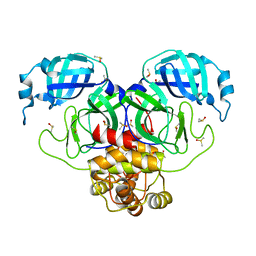 | | SARS-CoV-2 Main protease in orthorhombic space group | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Fearon, D, Douangamath, A, von Delft, F, Noske, G.D, Nakamura, A.M, Gawriljuk, V.O, Lima, G.M.A, Zeri, A.C.M, Nascimento, A.F.Z, Oliva, G, Godoy, A.S. | | Deposit date: | 2021-03-31 | | Release date: | 2021-04-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | A Crystallographic Snapshot of SARS-CoV-2 Main Protease Maturation Process.
J.Mol.Biol., 433, 2021
|
|
7GCF
 
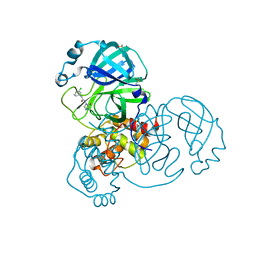 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with JAN-GHE-5a013bed-4 (Mpro-x10604) | | Descriptor: | 2-(3-chlorophenyl)-N-(5-oxo-1,5-dihydro-4H-1,2,4-triazol-4-yl)acetamide, 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-08 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.419 Å) | | Cite: | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
7GB1
 
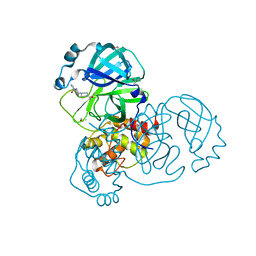 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with STE-KUL-2e0d2e88-2 (Mpro-x10150) | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE, N-[2-(4-acetylpiperazin-1-yl)ethyl]naphthalene-1-carboxamide | | Authors: | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-08 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.289 Å) | | Cite: | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
