7QC6
 
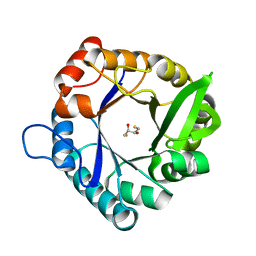 | | HisF_C9A_L50H_I52H mutant (apo) from T. maritima | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, Imidazole glycerol phosphate synthase subunit HisF | | Authors: | Beaumet, M, Dose, A, Braeuer, A, Mahy, J, Ghattas, W, Groll, M, Hess, C. | | Deposit date: | 2021-11-22 | | Release date: | 2022-08-10 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | An artificial metalloprotein with metal-adaptive coordination sites and Ni-dependent quercetinase activity.
J.Inorg.Biochem., 235, 2022
|
|
7QC7
 
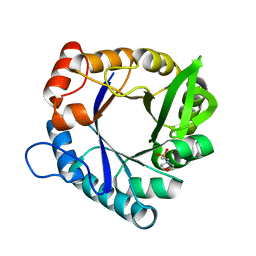 | | HisF-C9A-D11E-V33A_L50H_I52H mutant (apo) from T. maritima | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Imidazole glycerol phosphate synthase subunit HisF | | Authors: | Beaumet, M, Dose, A, Braeuer, A, Mahy, J, Ghattas, W, Groll, M, Hess, C. | | Deposit date: | 2021-11-22 | | Release date: | 2022-08-10 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | An artificial metalloprotein with metal-adaptive coordination sites and Ni-dependent quercetinase activity.
J.Inorg.Biochem., 235, 2022
|
|
7QC8
 
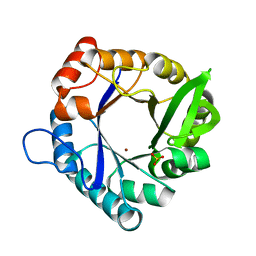 | | HisF-C9A-D11E-V33A_L50H_I52H mutant in complex with Zn(II) from T. maritima | | Descriptor: | Imidazole glycerol phosphate synthase subunit HisF, SULFATE ION, ZINC ION | | Authors: | Beaumet, M, Dose, A, Braeuer, A, Mahy, J, Ghattas, W, Groll, M, Hess, C. | | Deposit date: | 2021-11-22 | | Release date: | 2022-08-10 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | An artificial metalloprotein with metal-adaptive coordination sites and Ni-dependent quercetinase activity.
J.Inorg.Biochem., 235, 2022
|
|
7QC9
 
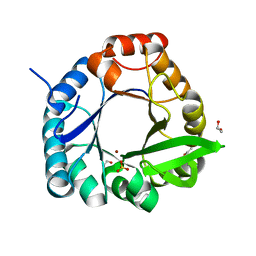 | | HisF-C9A-D11E-V33A_L50H_I52H mutant in complex with Ni(II) from T. maritima | | Descriptor: | 1,2-ETHANEDIOL, Imidazole glycerol phosphate synthase subunit HisF, NICKEL (II) ION, ... | | Authors: | Beaumet, M, Dose, A, Braeuer, A, Mahy, J, Ghattas, W, Groll, M, Hess, C. | | Deposit date: | 2021-11-22 | | Release date: | 2022-08-10 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | An artificial metalloprotein with metal-adaptive coordination sites and Ni-dependent quercetinase activity.
J.Inorg.Biochem., 235, 2022
|
|
7QC3
 
 | | HisF from T. maritima | | Descriptor: | Imidazole glycerol phosphate synthase subunit HisF | | Authors: | Beaumet, M, Dose, A, Braeuer, A, Mahy, J, Ghattas, W, Groll, M, Hess, C. | | Deposit date: | 2021-11-22 | | Release date: | 2022-08-10 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | An artificial metalloprotein with metal-adaptive coordination sites and Ni-dependent quercetinase activity.
J.Inorg.Biochem., 235, 2022
|
|
7U0L
 
 | | Crystal structure of the CCoV-HuPn-2018 RBD (domain B) in complex with canine APN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Aminopeptidase N, ... | | Authors: | Tortorici, M.A, Veesler, D, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2022-02-18 | | Release date: | 2022-08-24 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structure, receptor recognition, and antigenicity of the human coronavirus CCoV-HuPn-2018 spike glycoprotein.
Cell, 185, 2022
|
|
7USB
 
 | | CCoV-HuPn-2018 S in the swung out conformation (local refinement of domain 0) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Tortorici, M.A, Veesler, D, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2022-04-23 | | Release date: | 2022-08-24 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structure, receptor recognition, and antigenicity of the human coronavirus CCoV-HuPn-2018 spike glycoprotein.
Cell, 185, 2022
|
|
7USA
 
 | | Structure of the human coronavirus CCoV-HuPn-2018 spike glycoprotein with domain 0 in the swung out conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Tortorici, M.A, Veesler, D, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2022-04-23 | | Release date: | 2022-08-24 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structure, receptor recognition, and antigenicity of the human coronavirus CCoV-HuPn-2018 spike glycoprotein.
Cell, 185, 2022
|
|
8UR5
 
 | |
8UR7
 
 | |
7US9
 
 | | CCoV-HuPn-2018 S in the proximal conformation (local refinement of domain 0) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Tortorici, M.A, Veesler, D, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2022-04-23 | | Release date: | 2022-08-24 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structure, receptor recognition, and antigenicity of the human coronavirus CCoV-HuPn-2018 spike glycoprotein.
Cell, 185, 2022
|
|
7US6
 
 | | Structure of the human coronavirus CCoV-HuPn-2018 spike glycoprotein with domain 0 in the proximal conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Tortorici, M.A, Veesler, D, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2022-04-23 | | Release date: | 2022-08-24 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structure, receptor recognition, and antigenicity of the human coronavirus CCoV-HuPn-2018 spike glycoprotein.
Cell, 185, 2022
|
|
8F54
 
 | |
8F4X
 
 | |
8F53
 
 | |
9DND
 
 | | Pseudosymmetric protein nanocage GI4 -F7 (local refinement) | | Descriptor: | Pseudosymmetric protein nanocages GI4 A Chain, Pseudosymmetric protein nanocages GI4 B Chain, Pseudosymmetric protein nanocages GI4 C Chain | | Authors: | Park, Y.J, Dowling, Q.M, Seattle Structural Genomics Center for Infectious Disease (SSGCID), King, N.P, Veesler, D. | | Deposit date: | 2024-09-17 | | Release date: | 2024-12-18 | | Last modified: | 2025-02-26 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Hierarchical design of pseudosymmetric protein nanocages.
Nature, 638, 2025
|
|
9DNE
 
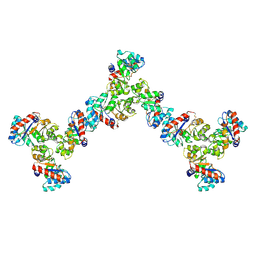 | | Pseudosymmetric protein nanocage GI9-F7 (local refinement) | | Descriptor: | Pseudosymmetric protein nanocage GI9-F7 A chain, Pseudosymmetric protein nanocage GI9-F7 B chain, Pseudosymmetric protein nanocage GI9-F7 C chain | | Authors: | Park, Y.J, Dowling, Q.M, King, N.P, Seattle Structural Genomics Center for Infectious Disease (SSGCID), Veesler, D. | | Deposit date: | 2024-09-17 | | Release date: | 2024-12-18 | | Last modified: | 2025-02-26 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Hierarchical design of pseudosymmetric protein nanocages.
Nature, 638, 2025
|
|
