1BEN
 
 | | INSULIN COMPLEXED WITH 4-HYDROXYBENZAMIDE | | 分子名称: | 4-HYDROXYBENZAMIDE, CHLORIDE ION, HUMAN INSULIN, ... | | 著者 | Smith, G.D, Ciszak, E, Pangborn, W. | | 登録日 | 1996-02-15 | | 公開日 | 1996-07-11 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | A novel complex of a phenolic derivative with insulin: structural features related to the T-->R transition.
Protein Sci., 5, 1996
|
|
1DPH
 
 | | CONFORMATIONAL CHANGES IN CUBIC INSULIN CRYSTALS IN THE PH RANGE 7-11 | | 分子名称: | 1,2-DICHLOROETHANE, INSULIN A CHAIN (PH 11), INSULIN B CHAIN (PH 11), ... | | 著者 | Gursky, O, Badger, J, Li, Y, Caspar, D.L.D. | | 登録日 | 1992-10-30 | | 公開日 | 1993-01-15 | | 最終更新日 | 2017-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Conformational changes in cubic insulin crystals in the pH range 7-11.
Biophys.J., 63, 1992
|
|
3ENG
 
 | | STRUCTURE OF ENDOGLUCANASE V CELLOBIOSE COMPLEX | | 分子名称: | ENDOGLUCANASE V CELLOBIOSE COMPLEX, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | 著者 | Davies, G.J, Schulein, M. | | 登録日 | 1996-10-17 | | 公開日 | 1997-06-16 | | 最終更新日 | 2023-08-09 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structure determination and refinement of the Humicola insolens endoglucanase V at 1.5 A resolution.
Acta Crystallogr.,Sect.D, 52, 1996
|
|
1MNI
 
 | |
1TGL
 
 | | A SERINE PROTEASE TRIAD FORMS THE CATALYTIC CENTRE OF A TRIACYLGLYCEROL LIPASE | | 分子名称: | TRIACYL-GLYCEROL ACYLHYDROLASE | | 著者 | Brady, L, Brzozowski, A.M, Derewenda, Z.S, Dodson, E.J, Dodson, G.G, Tolley, S.P, Turkenburg, J.P, Christiansen, L, Huge-Jensen, B, Norskov, L, Thim, L. | | 登録日 | 1990-02-05 | | 公開日 | 1990-10-15 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | A serine protease triad forms the catalytic centre of a triacylglycerol lipase.
Nature, 343, 1990
|
|
1RKM
 
 | | STRUCTURE OF OPPA | | 分子名称: | OLIGO-PEPTIDE BINDING PROTEIN | | 著者 | Sleigh, S.H, Tame, J.R.H, Wilkinson, A.J. | | 登録日 | 1997-03-25 | | 公開日 | 1997-07-29 | | 最終更新日 | 2023-08-09 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Peptide binding in OppA, the crystal structures of the periplasmic oligopeptide binding protein in the unliganded form and in complex with lysyllysine.
Biochemistry, 36, 1997
|
|
1APH
 
 | | CONFORMATIONAL CHANGES IN CUBIC INSULIN CRYSTALS IN THE PH RANGE 7-11 | | 分子名称: | 1,2-DICHLOROETHANE, INSULIN A CHAIN (PH 7), INSULIN B CHAIN (PH 7) | | 著者 | Gursky, O, Badger, J, Li, Y, Caspar, D.L.D. | | 登録日 | 1992-10-30 | | 公開日 | 1993-01-15 | | 最終更新日 | 2017-11-29 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Conformational changes in cubic insulin crystals in the pH range 7-11.
Biophys.J., 63, 1992
|
|
1BPH
 
 | | CONFORMATIONAL CHANGES IN CUBIC INSULIN CRYSTALS IN THE PH RANGE 7-11 | | 分子名称: | 1,2-DICHLOROETHANE, INSULIN A CHAIN (PH 9), INSULIN B CHAIN (PH 9), ... | | 著者 | Gursky, O, Badger, J, Li, Y, Caspar, D.L.D. | | 登録日 | 1992-10-30 | | 公開日 | 1993-01-15 | | 最終更新日 | 2017-11-29 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Conformational changes in cubic insulin crystals in the pH range 7-11.
Biophys.J., 63, 1992
|
|
1OLC
 
 | |
1PNK
 
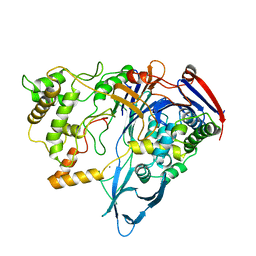 | |
1PNM
 
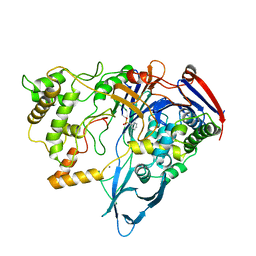 | |
1PNL
 
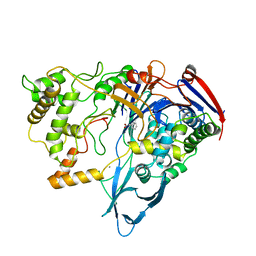 | |
2TPL
 
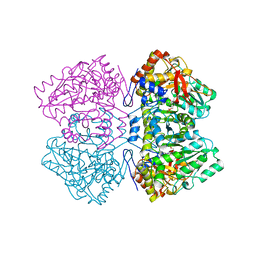 | | TYROSINE PHENOL-LYASE FROM CITROBACTER INTERMEDIUS COMPLEX WITH 3-(4'-HYDROXYPHENYL)PROPIONIC ACID, PYRIDOXAL-5'-PHOSPHATE AND CS+ ION | | 分子名称: | CESIUM ION, HYDROXYPHENYL PROPIONIC ACID, TYROSINE PHENOL-LYASE | | 著者 | Antson, A.A, Demidkina, T.V, Wilson, K.S. | | 登録日 | 1997-01-23 | | 公開日 | 1997-04-01 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | The crystal structure of Citrobacter freundii tyrosine phenol-lyase complexed with 3-(4'-hydroxyphenyl)propionic acid, together with site-directed mutagenesis and kinetic analysis, demonstrates that arginine 381 is required for substrate specificity.
Biochemistry, 36, 1997
|
|
2RJI
 
 | |
2RBI
 
 | |
1TYM
 
 | |
1UTD
 
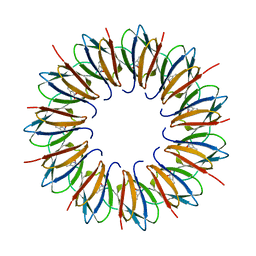 | | The structure of the trp RNA-binding attenuation protein (TRAP) bound to a 63-nucleotide RNA molecule containing GAGUUU repeats | | 分子名称: | 5'-R(*GP*UP*UP*UP*GP*AP)-3', TRANSCRIPTION ATTENUATION PROTEIN MTRB, TRYPTOPHAN | | 著者 | Hopcroft, N.H, Manfredo, A, Wendt, A.L, Brzozowski, A.M, Gollnick, P, Antson, A.A. | | 登録日 | 2003-12-08 | | 公開日 | 2004-01-15 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | The Interaction of RNA with Trap: The Role of Triplet Repeats and Separating Spacer Nucleotides
J.Mol.Biol., 338, 2004
|
|
1EV3
 
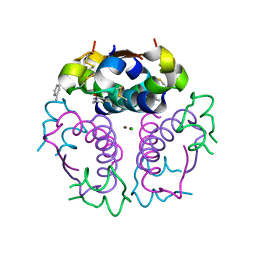 | | Structure of the rhombohedral form of the M-cresol/insulin R6 hexamer | | 分子名称: | CHLORIDE ION, INSULIN, M-CRESOL, ... | | 著者 | Smith, G.D, Ciszak, E, Magrum, L.A, Pangborn, W.A, Blessing, R.H. | | 登録日 | 2000-04-19 | | 公開日 | 2000-12-04 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (1.78 Å) | | 主引用文献 | R6 hexameric insulin complexed with m-cresol or resorcinol.
Acta Crystallogr.,Sect.D, 56, 2000
|
|
1EV6
 
 | | Structure of the monoclinic form of the M-cresol/insulin R6 hexamer | | 分子名称: | CHLORIDE ION, INSULIN, M-CRESOL, ... | | 著者 | Smith, G.D, Ciszak, E, Magrum, L.A, Pangborn, W.A, Blessing, R.H. | | 登録日 | 2000-04-19 | | 公開日 | 2000-12-04 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | R6 Hexameric Insulin Complexed with m-Cresol or Resorcinol
Biochem.Biophys.Res.Commun., 56, 2000
|
|
1EVR
 
 | | The structure of the resorcinol/insulin R6 hexamer | | 分子名称: | CHLORIDE ION, INSULIN, RESORCINOL, ... | | 著者 | Smith, G.D, Ciszak, E, Magrum, L.A, Pangborn, W.A, Blessing, R.H. | | 登録日 | 2000-04-20 | | 公開日 | 2000-12-04 | | 最終更新日 | 2017-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | R6 hexameric insulin complexed with m-cresol or resorcinol.
Acta Crystallogr.,Sect.D, 56, 2000
|
|
1QKA
 
 | |
1QKB
 
 | | OLIGO-PEPTIDE BINDING PROTEIN (OPPA) COMPLEXED WITH KVK | | 分子名称: | ACETATE ION, PEPTIDE LYS-VAL-LYS, PERIPLASMIC OLIGOPEPTIDE-BINDING PROTEIN, ... | | 著者 | Tame, J.R.H, Sleigh, S.H, Wilkinson, A.J. | | 登録日 | 1999-07-14 | | 公開日 | 1999-09-09 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Crystallographic and Calorimetric Analysis of Peptide Binding to Oppa Protein
J.Mol.Biol., 291, 1999
|
|
1LPH
 
 | | LYS(B28)PRO(B29)-HUMAN INSULIN | | 分子名称: | CHLORIDE ION, INSULIN, PHENOL, ... | | 著者 | Ciszak, E, Beals, J.M, Frank, B.H, Baker, J.C, Carter, N.D, Smith, G.D. | | 登録日 | 1995-04-19 | | 公開日 | 1996-06-20 | | 最終更新日 | 2021-11-03 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Role of C-terminal B-chain residues in insulin assembly: the structure of hexameric LysB28ProB29-human insulin.
Structure, 3, 1995
|
|
1MNH
 
 | | INTERACTIONS AMONG RESIDUES CD3, E7, E10 AND E11 IN MYOGLOBINS: ATTEMPTS TO SIMULATE THE O2 AND CO BINDING PROPERTIES OF APLYSIA MYOGLOBIN | | 分子名称: | MYOGLOBIN, PROTOPORPHYRIN IX CONTAINING FE | | 著者 | Davies, G.J, Wilkinson, A.J. | | 登録日 | 1995-01-11 | | 公開日 | 1995-05-08 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Interactions among residues CD3, E7, E10, and E11 in myoglobins: attempts to simulate the ligand-binding properties of Aplysia myoglobin.
Biochemistry, 34, 1995
|
|
1MNK
 
 | | INTERACTIONS AMONG RESIDUES CD3, E7, E10 AND E11 IN MYOGLOBINS: ATTEMPTS TO SIMULATE THE O2 AND CO BINDING PROPERTIES OF APLYSIA MYOGLOBIN | | 分子名称: | MYOGLOBIN, PROTOPORPHYRIN IX CONTAINING FE | | 著者 | Krzywda, S, Wilkinson, A.J. | | 登録日 | 1995-01-11 | | 公開日 | 1995-04-20 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Interactions among residues CD3, E7, E10, and E11 in myoglobins: attempts to simulate the ligand-binding properties of Aplysia myoglobin.
Biochemistry, 34, 1995
|
|
