1DGN
 
 | | SOLUTION STRUCTURE OF ICEBERG, AN INHIBITOR OF INTERLEUKIN-1BETA GENERATION | | Descriptor: | ICEBERG (PROTEASE INHIBITOR) | | Authors: | Humke, E.W, Shriver, S.K, Starovasnik, M.A, Fairbrother, W.J, Dixit, V.M. | | Deposit date: | 1999-11-24 | | Release date: | 2000-10-09 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | ICEBERG: a novel inhibitor of interleukin-1beta generation.
Cell(Cambridge,Mass.), 103, 2000
|
|
1RJ8
 
 | | The crystal structure of TNF family member EDA-A2 | | Descriptor: | ectodysplasin-A isoform EDA-A2 | | Authors: | Hymowitz, S.G, Compaan, D.M, Yan, M, Ackerly, H, Dixit, V.M, Starovasnik, M.A, de Vos, A.M. | | Deposit date: | 2003-11-18 | | Release date: | 2003-12-09 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | The Crystal Structure of EDA-A1 and EDA-A2: splice variants with distinct receptor specificity
STRUCTURE, 11, 2003
|
|
1RJ7
 
 | | Crystal structure of EDA-A1 | | Descriptor: | Ectodysplasin A | | Authors: | Hymowitz, S.G, Compaan, D.M, Yan, M, Ackerly, H, Dixit, V.M, Starovasnik, M.A, de Vos, A.M. | | Deposit date: | 2003-11-18 | | Release date: | 2003-12-09 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The crystal structures of EDA-A1 and EDA-A2: splice variants with distinct receptor specificity.
Structure, 11, 2003
|
|
3U30
 
 | | Crystal structure of a linear-specific Ubiquitin fab bound to linear ubiquitin | | Descriptor: | Heavy chain Fab, Light chain Fab, linear di-ubiquitin | | Authors: | Matsumoto, M.L, Dong, K.C, Yu, C, Phu, L, Gao, X, Hannoush, R.N, Hymowitz, S.G, Kirkpatrick, D.S, Dixit, V.M, Kelley, R.F. | | Deposit date: | 2011-10-04 | | Release date: | 2012-01-18 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.428 Å) | | Cite: | Engineering and structural characterization of a linear polyubiquitin-specific antibody.
J.Mol.Biol., 418, 2012
|
|
1OSG
 
 | | Complex between BAFF and a BR3 derived peptide presented in a beta-hairpin scaffold | | Descriptor: | BR3 derived PEPTIDE, MAGNESIUM ION, Tumor necrosis factor ligand superfamily member 13B | | Authors: | Gordon, N.C, Pan, B, Hymowitz, S.G, Yin, J.P, Kelley, R.F, Cochran, A.G, Yan, M, Dixit, V.M, Fairbrother, W.J, Starovasnik, M.A. | | Deposit date: | 2003-03-19 | | Release date: | 2003-05-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | BAFF/BLyS receptor 3 comprises a minimal TNF receptor-like module that encodes a highly focused ligand-binding site
Biochemistry, 42, 2003
|
|
1OSX
 
 | | Solution Structure of the Extracellular Domain of BLyS Receptor 3 (BR3) | | Descriptor: | Tumor necrosis factor receptor superfamily member 13C | | Authors: | Gordon, N.C, Pan, B, Hymowitz, S.G, Yin, J.P, Kelley, R.F, Cochran, A.G, Yan, M, Dixit, V.M, Fairbrother, W.J, Starovasnik, M.A. | | Deposit date: | 2003-03-20 | | Release date: | 2003-05-27 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | BAFF/BLyS receptor 3 comprises a minimal TNF receptor-like module that encodes a highly focused ligand-binding site
Biochemistry, 42, 2003
|
|
1MPV
 
 | | Structure of bhpBR3, the BAFF-binding loop of BR3 embedded in a beta-hairpin peptide | | Descriptor: | BLyS Receptor 3 | | Authors: | Kayagaki, N, Yan, M, Seshasayee, D, Wang, H, Lee, W, French, D.M, Grewal, I.S, Cochran, A.G, Gordon, N.C, Yin, J, Starovasnik, M.A, Dixit, V.M. | | Deposit date: | 2002-09-12 | | Release date: | 2002-10-30 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | BAFF/BLyS receptor 3 binds the B cell survival factor BAFF ligand through a discrete surface loop and promotes processing of NF-kappaB2.
Immunity, 17, 2002
|
|
9BIA
 
 | |
1OY7
 
 | | Structure and Function Analysis of Peptide Antagonists of Melanoma Inhibitor of Apoptosis (ML-IAP) | | Descriptor: | 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, AEVVAVKSE peptide, Baculoviral IAP repeat-containing protein 7, ... | | Authors: | Franklin, M.C, Kadkhodayan, S, Ackerly, H, Alexandru, D, Distefano, M.D, Elliott, L.O, Flygare, J.A, Vucic, D, Deshayes, K, Fairbrother, W.J. | | Deposit date: | 2003-04-03 | | Release date: | 2003-08-26 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure and Function Analysis of Peptide Antagonists of Melanoma Inhibitor of Apoptosis (ML-IAP)
Biochemistry, 42, 2003
|
|
1OXN
 
 | | Structure and Function Analysis of Peptide Antagonists of Melanoma Inhibitor of Apoptosis (ML-IAP) | | Descriptor: | 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, AEAVPWKSE peptide, Baculoviral IAP repeat-containing protein 7, ... | | Authors: | Franklin, M.C, Kadkhodayan, S, Ackerly, H, Alexandru, D, Distefano, M.D, Elliott, L.O, Flygare, J.A, Vucic, D, Deshayes, K, Fairbrother, W.J. | | Deposit date: | 2003-04-03 | | Release date: | 2003-08-26 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure and Function Analysis of Peptide Antagonists of Melanoma Inhibitor of Apoptosis (ML-IAP)
Biochemistry, 42, 2003
|
|
1OXQ
 
 | | Structure and Function Analysis of Peptide Antagonists of Melanoma Inhibitor of Apoptosis (ML-IAP) | | Descriptor: | 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, AVPIAQKSE (Smac) peptide, Baculoviral IAP repeat-containing protein 7, ... | | Authors: | Franklin, M.C, Kadkhodayan, S, Ackerly, H, Alexandru, D, Distefano, M.D, Elliott, L.O, Flygare, J.A, Vucic, D, Deshayes, K, Fairbrother, W.J. | | Deposit date: | 2003-04-03 | | Release date: | 2003-08-26 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure and Function Analysis of Peptide Antagonists of Melanoma Inhibitor of Apoptosis (ML-IAP)
Biochemistry, 42, 2003
|
|
9DHD
 
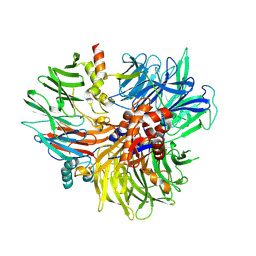 | | The ternary complex of DDB1, DDA1, DET1 | | Descriptor: | DET1 homolog, DET1- and DDB1-associated protein 1, DNA damage-binding protein 1 | | Authors: | Schubert, A.F, Kschonsak, M, Harris, S.F. | | Deposit date: | 2024-09-03 | | Release date: | 2025-02-05 | | Last modified: | 2025-03-05 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Impairment of DET1 causes neurological defects and lethality in mice and humans.
Proc.Natl.Acad.Sci.USA, 122, 2025
|
|
8T48
 
 | |
7LW1
 
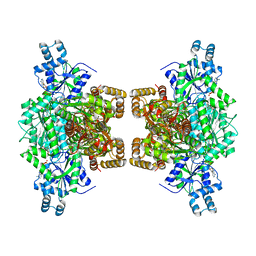 | | Human phosphofructokinase-1 liver type bound to activator NA-11 | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, 6-O-phosphono-beta-D-fructofuranose, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Lynch, E.M, Kollman, J.M, Webb, B. | | Deposit date: | 2021-02-27 | | Release date: | 2022-01-26 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Selective activation of PFKL suppresses the phagocytic oxidative burst.
Cell, 184, 2021
|
|
3NOB
 
 | |
3OJ4
 
 | |
3OJ3
 
 | |
3PTF
 
 | |
5DS8
 
 | |
5DTF
 
 | |
5DSC
 
 | |
5DUB
 
 | |
3DVN
 
 | |
5DRN
 
 | |
3DVG
 
 | |
