5TB3
 
 | | Structure of rabbit RyR1 (EGTA-only dataset, class 3) | | Descriptor: | Peptidyl-prolyl cis-trans isomerase FKBP1B, Ryanodine receptor 1, ZINC ION | | Authors: | Clarke, O.B, des Georges, A, Zalk, R, Marks, A.R, Hendrickson, W.A, Frank, J. | | Deposit date: | 2016-09-11 | | Release date: | 2016-10-12 | | Last modified: | 2018-07-18 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | Structural Basis for Gating and Activation of RyR1.
Cell, 167, 2016
|
|
5T9V
 
 | | Structure of rabbit RyR1 (Caffeine/ATP/Ca2+ dataset, class 1) | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CAFFEINE, CALCIUM ION, ... | | Authors: | Clarke, O.B, des Georges, A, Zalk, R, Marks, A.R, Hendrickson, W.A, Frank, J. | | Deposit date: | 2016-09-09 | | Release date: | 2016-10-12 | | Last modified: | 2018-07-18 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Structural Basis for Gating and Activation of RyR1.
Cell, 167, 2016
|
|
5TAX
 
 | | Structure of rabbit RyR1 (ryanodine dataset, class 1) | | Descriptor: | CALCIUM ION, Peptidyl-prolyl cis-trans isomerase FKBP1B, Ryanodine receptor 1, ... | | Authors: | Clarke, O.B, des Georges, A, Zalk, R, Marks, A.R, Hendrickson, W.A, Frank, J. | | Deposit date: | 2016-09-10 | | Release date: | 2016-10-12 | | Last modified: | 2018-07-18 | | Method: | ELECTRON MICROSCOPY (6.6 Å) | | Cite: | Structural Basis for Gating and Activation of RyR1.
Cell, 167, 2016
|
|
6PV6
 
 | | Functional Pathways of Biomolecules Retrieved from Single-particle Snapshots | | Descriptor: | CALCIUM ION, Peptidyl-prolyl cis-trans isomerase FKBP1B, Ryanodine receptor 1, ... | | Authors: | Dashti, A, des Georges, A, Frank, J, Ourmazd, A. | | Deposit date: | 2019-07-19 | | Release date: | 2020-08-12 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Retrieving functional pathways of biomolecules from single-particle snapshots.
Nat Commun, 11, 2020
|
|
4V8M
 
 | | High-resolution cryo-electron microscopy structure of the Trypanosoma brucei ribosome | | Descriptor: | 18S RRNA OF THE SMALL RIBOSOMAL SUBUNIT, 40S RIBOSOMAL PROTEIN S10, PUTATIVE, ... | | Authors: | Hashem, Y, des Georges, A, Fu, J, Buss, S.N, Jossinet, F, Jobe, A, Zhang, Q, Liao, H.Y, Grassucci, R.A, Bajaj, C, Westhof, E, Madison-Antenucci, S, Frank, J. | | Deposit date: | 2012-12-09 | | Release date: | 2014-07-09 | | Last modified: | 2019-12-11 | | Method: | ELECTRON MICROSCOPY (5.57 Å) | | Cite: | High-Resolution Cryo-Electron Microscopy Structure of the Trypanosoma Brucei Ribosome.
Nature, 494, 2013
|
|
6GSR
 
 | | Single Particle Cryo-EM map of human Transferrin receptor 1 - H-Ferritin complex at 5.5 Angstrom resolution. | | Descriptor: | Ferritin heavy chain, Transferrin receptor protein 1 | | Authors: | Testi, C, Montemiglio, L.C, Vallone, B, Des Georges, A, Boffi, A, Mancia, F, Baiocco, P. | | Deposit date: | 2018-06-15 | | Release date: | 2019-03-27 | | Last modified: | 2019-05-08 | | Method: | ELECTRON MICROSCOPY (5.5 Å) | | Cite: | Cryo-EM structure of the human ferritin-transferrin receptor 1 complex.
Nat Commun, 10, 2019
|
|
6H5I
 
 | | Single Particle Cryo-EM map of human Transferrin receptor 1 - H-Ferritin complex. | | Descriptor: | Ferritin heavy chain, Transferrin receptor protein 1 | | Authors: | Testi, C, Montemiglio, L.C, Vallone, B, Des Georges, A, Boffi, A, Mancia, F, Baiocco, P, Savino, C. | | Deposit date: | 2018-07-24 | | Release date: | 2019-03-27 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Cryo-EM structure of the human ferritin-transferrin receptor 1 complex.
Nat Commun, 10, 2019
|
|
7ZQS
 
 | | Cryo-EM Structure of Human Transferrin Receptor 1 bound to DNA Aptamer | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, DNA (30-MER), ... | | Authors: | Bansia, H, Wang, T, Gutierrez, D, des Georges, A. | | Deposit date: | 2022-05-02 | | Release date: | 2022-08-17 | | Method: | ELECTRON MICROSCOPY (2.54 Å) | | Cite: | Discovery of a Transferrin Receptor 1-Binding Aptamer and Its Application in Cancer Cell Depletion for Adoptive T-Cell Therapy Manufacturing.
J.Am.Chem.Soc., 144, 2022
|
|
6NI2
 
 | | Stabilized beta-arrestin 1-V2T subcomplex of a GPCR-G protein-beta-arrestin mega-complex | | Descriptor: | Beta-arrestin-1, Fab30 Heavy Chain, Fab30 Light Chain, ... | | Authors: | Nguyen, A.H, Thomsen, A.R.B, Cahill, T.J, des Georges, A, Lefkowitz, R.J. | | Deposit date: | 2018-12-26 | | Release date: | 2019-11-20 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structure of an endosomal signaling GPCR-G protein-beta-arrestin megacomplex.
Nat.Struct.Mol.Biol., 26, 2019
|
|
7N4T
 
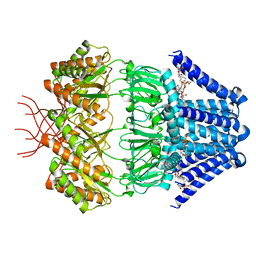 | | Low conductance mechanosensitive channel YnaI | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, Low conductance mechanosensitive channel YnaI | | Authors: | Catalano, C, Ben-Hail, D, Qiu, W, des Georges, A, Guo, Y. | | Deposit date: | 2021-06-04 | | Release date: | 2022-04-20 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | Cryo-EM Structure of Mechanosensitive Channel YnaI Using SMA2000: Challenges and Opportunities.
Membranes (Basel), 11, 2021
|
|
6NI3
 
 | | B2V2R-Gs protein subcomplex of a GPCR-G protein-beta-arrestin mega-complex | | Descriptor: | 8-[(1R)-2-{[1,1-dimethyl-2-(2-methylphenyl)ethyl]amino}-1-hydroxyethyl]-5-hydroxy-2H-1,4-benzoxazin-3(4H)-one, Endolysin,Beta-2 adrenergic receptor chimera, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Nguyen, A.H, Thomsen, A.R.B, Cahill, T.J, des Georges, A, Lefkowitz, R.J. | | Deposit date: | 2018-12-26 | | Release date: | 2019-11-20 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structure of an endosomal signaling GPCR-G protein-beta-arrestin megacomplex.
Nat.Struct.Mol.Biol., 26, 2019
|
|
7RR7
 
 | | Multidrug efflux pump subunit AcrB | | Descriptor: | DODECANE, Multidrug efflux pump subunit AcrB, PHOSPHATIDYLETHANOLAMINE | | Authors: | Trinh, T.K.H, Cabezas, A, Catalano, C, Qiu, W, des Georges, A, Guo, Y. | | Deposit date: | 2021-08-09 | | Release date: | 2022-08-17 | | Method: | ELECTRON MICROSCOPY (3.05 Å) | | Cite: | Multidrug efflux pump subunit AcrB
To Be Published
|
|
7RR8
 
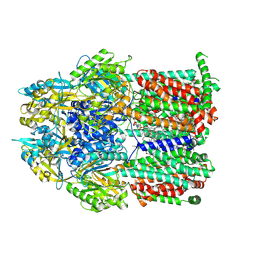 | | Multidrug efflux pump subunit AcrB | | Descriptor: | DODECANE, Multidrug efflux pump subunit AcrB, PHOSPHATIDYLETHANOLAMINE | | Authors: | Trinh, T.K.H, Cabezas, A, Catalano, C, Qiu, W, des Georges, A, Guo, Y. | | Deposit date: | 2021-08-09 | | Release date: | 2022-08-17 | | Method: | ELECTRON MICROSCOPY (3.51 Å) | | Cite: | Multidrug efflux pump subunit AcrB
To Be Published
|
|
7RR6
 
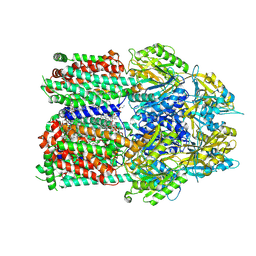 | | Multidrug efflux pump subunit AcrB | | Descriptor: | DODECANE, Multidrug efflux pump subunit AcrB, PHOSPHATIDYLETHANOLAMINE | | Authors: | Trinh, T.K.H, Cabezas, A, Catalano, C, Qiu, W, des Georges, A, Guo, Y. | | Deposit date: | 2021-08-09 | | Release date: | 2022-08-17 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Multidrug efflux pump subunit AcrB
To Be Published
|
|
7RAZ
 
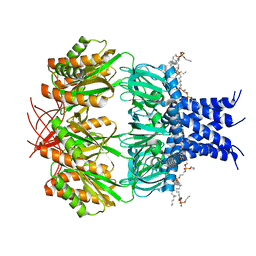 | | Small conductance mechanosensitive channel MscS | | Descriptor: | PHOSPHATIDYLETHANOLAMINE, Small-conductance mechanosensitive channel | | Authors: | Catalano, C, Ben-Hail, D, Qiu, W, des Georges, A, Guo, Y. | | Deposit date: | 2021-07-05 | | Release date: | 2022-07-13 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Small conductance mechanosensitive channel MscS
To Be Published
|
|
6BBM
 
 | | Mechanisms of Opening and Closing of the Bacterial Replicative Helicase: The DnaB Helicase and Lambda P Helicase Loader Complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Replication protein P, Replicative DNA helicase | | Authors: | Chase, J, Catalano, A, Noble, A.J, Eng, E.T, Olinares, P.D.B, Molloy, K, Pakotiprapha, D, Samuels, M, Chain, B, des Georges, A, Jeruzalmi, D. | | Deposit date: | 2017-10-18 | | Release date: | 2019-03-06 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Mechanisms of opening and closing of the bacterial replicative helicase.
Elife, 7, 2018
|
|
7JMJ
 
 | | Functional Pathways of Biomolecules Retrieved from Single-particle Snapshots - Frame 37 - State 5 (S5) | | Descriptor: | CALCIUM ION, Peptidyl-prolyl cis-trans isomerase FKBP1B, ZINC ION, ... | | Authors: | Dashti, A, des Georges, A, Frank, J, Ourmazd, A. | | Deposit date: | 2020-07-31 | | Release date: | 2020-08-12 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Retrieving functional pathways of biomolecules from single-particle snapshots.
Nat Commun, 11, 2020
|
|
7JMI
 
 | | Functional Pathways of Biomolecules Retrieved from Single-particle Snapshots - Frame 29 - State 3 (S3) | | Descriptor: | CALCIUM ION, Peptidyl-prolyl cis-trans isomerase FKBP1B, ZINC ION, ... | | Authors: | Dashti, A, des Georges, A, Frank, J, Ourmazd, A. | | Deposit date: | 2020-07-31 | | Release date: | 2020-08-12 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Retrieving functional pathways of biomolecules from single-particle snapshots.
Nat Commun, 11, 2020
|
|
7JMG
 
 | | Functional Pathways of Biomolecules Retrieved from Single-particle Snapshots - Frame 22 - State 2 (S2) | | Descriptor: | CALCIUM ION, Peptidyl-prolyl cis-trans isomerase FKBP1B, ZINC ION, ... | | Authors: | Dashti, A, des Georges, A, Frank, J, Ourmazd, A. | | Deposit date: | 2020-07-31 | | Release date: | 2020-08-12 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Retrieving functional pathways of biomolecules from single-particle snapshots.
Nat Commun, 11, 2020
|
|
7JMF
 
 | | Functional Pathways of Biomolecules Retrieved from Single-particle Snapshots - Frame 42 - State 6 (S6) | | Descriptor: | CALCIUM ION, Peptidyl-prolyl cis-trans isomerase FKBP1B, ZINC ION, ... | | Authors: | Dashti, A, des Georges, A, Frank, J, Ourmazd, A. | | Deposit date: | 2020-07-31 | | Release date: | 2020-08-12 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Retrieving functional pathways of biomolecules from single-particle snapshots.
Nat Commun, 11, 2020
|
|
7JMH
 
 | | Functional Pathways of Biomolecules Retrieved from Single-particle Snapshots - Frame 35 - State 4 (S4) | | Descriptor: | CALCIUM ION, Peptidyl-prolyl cis-trans isomerase FKBP1B, ZINC ION, ... | | Authors: | Dashti, A, des Georges, A, Frank, J, Ourmazd, A. | | Deposit date: | 2020-07-31 | | Release date: | 2020-08-12 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Retrieving functional pathways of biomolecules from single-particle snapshots.
Nat Commun, 11, 2020
|
|
3J8B
 
 | | Model of the human eIF3 PCI-MPN octamer docked into the 43S-HCV IRES EM map | | Descriptor: | Eukaryotic translation initiation factor 3 subunit A, Eukaryotic translation initiation factor 3 subunit C, Eukaryotic translation initiation factor 3 subunit E, ... | | Authors: | Erzberger, J.P, Ban, N. | | Deposit date: | 2014-10-08 | | Release date: | 2014-10-22 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (9.3 Å) | | Cite: | Molecular Architecture of the 40SeIF1eIF3 Translation Initiation Complex.
Cell(Cambridge,Mass.), 158, 2014
|
|
3J8C
 
 | | Model of the human eIF3 PCI-MPN octamer docked into the 43S EM map | | Descriptor: | Eukaryotic translation initiation factor 3 subunit A, Eukaryotic translation initiation factor 3 subunit C, Eukaryotic translation initiation factor 3 subunit E, ... | | Authors: | Erzberger, J.P, Ban, N. | | Deposit date: | 2014-10-08 | | Release date: | 2014-10-22 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (11.6 Å) | | Cite: | Molecular Architecture of the 40SeIF1eIF3 Translation Initiation Complex.
Cell(Cambridge,Mass.), 158, 2014
|
|
6UWT
 
 | |
6UWR
 
 | |
