2GQC
 
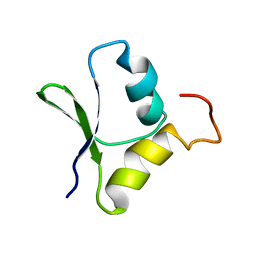 | | Solution structure of the N-terminal domain of Rhomboid Intramembrane Protease from P. aeruginosa | | Descriptor: | Rhomboid Intramembrane Protease | | Authors: | Dutta, K, Del Rio, A, Chavez, J, Ubarretxena-Belandia, I, Ghose, R. | | Deposit date: | 2006-04-20 | | Release date: | 2007-03-06 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure and dynamics of the N-terminal cytosolic domain of rhomboid intramembrane protease from Pseudomonas aeruginosa: insights into a functional role in intramembrane proteolysis.
J.Mol.Biol., 365, 2007
|
|
1AU7
 
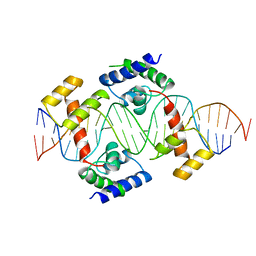 | | PIT-1 MUTANT/DNA COMPLEX | | Descriptor: | CONSENSUS DNA 25-MER, DNA (5'-D(*CP*TP*TP*CP*CP*TP*CP*AP*TP*GP*TP*AP*TP*AP*TP*AP*C P*AP*TP*GP*AP*GP* GP*A)-3'), PROTEIN PIT-1 | | Authors: | Jacobson, E.M, Li, P, Leon-Del-Rio, A, Rosenfeld, M.G, Aggarwal, A.K. | | Deposit date: | 1997-09-12 | | Release date: | 1998-01-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of Pit-1 POU domain bound to DNA as a dimer: unexpected arrangement and flexibility.
Genes Dev., 11, 1997
|
|
6ZRB
 
 | |
6YUH
 
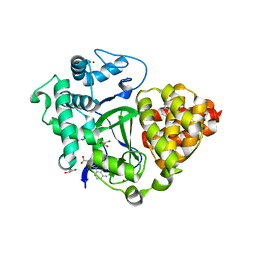 | | Crystal structure of SMYD3 with diperodon R enantiomer bound to allosteric site | | Descriptor: | Diperodon, GLYCEROL, Histone-lysine N-methyltransferase SMYD3, ... | | Authors: | Cederfelt, D, Talibov, V.O, Dobritzsch, D, Danielson, U.H. | | Deposit date: | 2020-04-27 | | Release date: | 2021-01-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Discovery of an Allosteric Ligand Binding Site in SMYD3 Lysine Methyltransferase.
Chembiochem, 22, 2021
|
|
8W2E
 
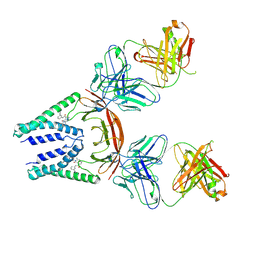 | | SARS-CoV-2 M protein dimer in complex with JNJ-9676 and Fab-B | | Descriptor: | (6S,8R)-N-(3-cyanophenyl)-5-{4-[difluoro(phenyl)methyl]phenyl}-6-methyl-4-oxo-4,5,6,7-tetrahydropyrazolo[1,5-a]pyrazine-3-carboxamide, Fab B Heavy Chain, Fab B Light Chain, ... | | Authors: | Yin, Y, Van Damme, E. | | Deposit date: | 2024-02-20 | | Release date: | 2025-01-29 | | Last modified: | 2025-04-23 | | Method: | ELECTRON MICROSCOPY (3.06 Å) | | Cite: | A small-molecule SARS-CoV-2 inhibitor targeting the membrane protein.
Nature, 640, 2025
|
|
6CZF
 
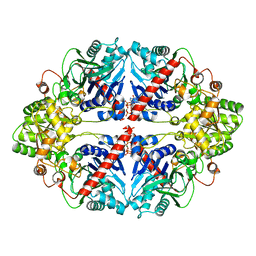 | | The structure of E. coli PurF in complex with ppGpp-Mg | | Descriptor: | Amidophosphoribosyltransferase, GUANOSINE-5',3'-TETRAPHOSPHATE, MAGNESIUM ION | | Authors: | Wang, B, Grant, R.A, Laub, M.T. | | Deposit date: | 2018-04-09 | | Release date: | 2018-10-10 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.949 Å) | | Cite: | Affinity-based capture and identification of protein effectors of the growth regulator ppGpp.
Nat. Chem. Biol., 15, 2019
|
|
