6HN6
 
 | | A revisited version of the apo structure of the ligand-binding domain of the human nuclear receptor RXR-ALPHA | | 分子名称: | 3-[(3-CHOLAMIDOPROPYL)DIMETHYLAMMONIO]-1-PROPANESULFONATE, Retinoic acid receptor RXR-alpha | | 著者 | Eberhardt, J, McEwen, A.G, Bourguet, W, Moras, D, Dejaegere, A. | | 登録日 | 2018-09-14 | | 公開日 | 2019-02-20 | | 最終更新日 | 2023-03-08 | | 実験手法 | X-RAY DIFFRACTION (2.71 Å) | | 主引用文献 | A revisited version of the apo structure of the ligand-binding domain of the human nuclear receptor retinoic X receptor alpha.
Acta Crystallogr F Struct Biol Commun, 75, 2019
|
|
5OEJ
 
 | | Structure of Tra1 subunit within the chromatin modifying complex SAGA | | 分子名称: | Tra1 subunit within the chromatin modifying complex SAGA | | 著者 | Sharov, G, Voltz, K, Durand, A, Kolesnikova, O, Papai, G, Myasnikov, A.G, Dejaegere, A, Ben-Shem, A, Schultz, P. | | 登録日 | 2017-07-07 | | 公開日 | 2017-08-02 | | 最終更新日 | 2017-11-29 | | 実験手法 | ELECTRON MICROSCOPY (5.7 Å) | | 主引用文献 | Structure of the transcription activator target Tra1 within the chromatin modifying complex SAGA.
Nat Commun, 8, 2017
|
|
1DW4
 
 | | NMR STRUCTURE OF OMEGA-CONOTOXIN MVIIA: CONSTRAINTS ON DISULPHIDE BRIDGES | | 分子名称: | OMEGA-CONOTOXIN MVIIA | | 著者 | Atkinson, R.A, Kieffer, B, Dejaegere, A, Sirockin, F, Lefevre, J.-F. | | 登録日 | 2000-01-24 | | 公開日 | 2000-03-01 | | 最終更新日 | 2022-02-16 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structural and dynamic characterization of omega-conotoxin MVIIA: the binding loop exhibits slow conformational exchange.
Biochemistry, 39, 2000
|
|
1DW5
 
 | | NMR STRUCTURE OF OMEGA-CONOTOXIN MVIIA: NO CONSTRAINTS ON DISULPHIDE BRIDGES | | 分子名称: | OMEGA-CONOTOXIN MVIIA | | 著者 | Atkinson, R.A, Kieffer, B, Dejaegere, A, Sirockin, F, Lefevre, J.-F. | | 登録日 | 2000-01-24 | | 公開日 | 2000-03-01 | | 最終更新日 | 2022-02-16 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structural and dynamic characterization of omega-conotoxin MVIIA: the binding loop exhibits slow conformational exchange.
Biochemistry, 39, 2000
|
|
3Q4J
 
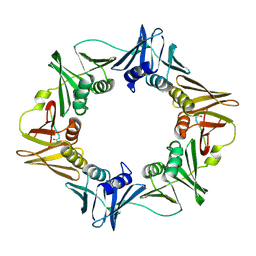 | | Structure of a small peptide ligand bound to E.coli DNA sliding clamp | | 分子名称: | DNA polymerase III subunit beta, peptide ligand | | 著者 | Wolff, P, Olieric, V, Briand, J.P, Chaloin, O, Dejaegere, A, Dumas, P, Ennifar, E, Guichard, G, Wagner, J, Burnouf, D. | | 登録日 | 2010-12-23 | | 公開日 | 2011-12-28 | | 最終更新日 | 2013-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structure-based design of short peptide ligands binding onto the E. coli processivity ring.
J.Med.Chem., 54, 2011
|
|
3Q4K
 
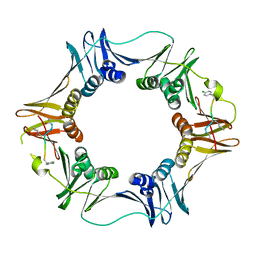 | | Structure of a small peptide ligand bound to E.coli DNA sliding clamp | | 分子名称: | DNA polymerase III subunit beta, peptide ligand | | 著者 | Wolff, P, Olieric, V, Briand, J.P, Chaloin, O, Dejaegere, A, Dumas, P, Ennifar, E, Guichard, G, Wagner, J, Burnouf, D. | | 登録日 | 2010-12-23 | | 公開日 | 2011-12-28 | | 最終更新日 | 2013-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Structure-based design of short peptide ligands binding onto the E. coli processivity ring.
J.Med.Chem., 54, 2011
|
|
3Q4L
 
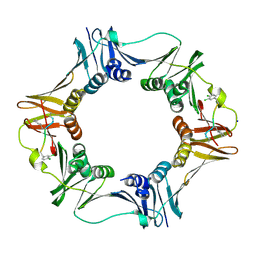 | | Structure of a small peptide ligand bound to E.coli DNA sliding clamp | | 分子名称: | DNA polymerase III subunit beta, SODIUM ION, peptide ligand | | 著者 | Wolff, P, Olieric, V, Briand, J.P, Chaloin, O, Dejaegere, A, Dumas, P, Ennifar, E, Guichard, G, Wagner, J, Burnouf, D. | | 登録日 | 2010-12-23 | | 公開日 | 2011-12-28 | | 最終更新日 | 2013-03-20 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Structure-based design of short peptide ligands binding onto the E. coli processivity ring.
J.Med.Chem., 54, 2011
|
|
4Q0A
 
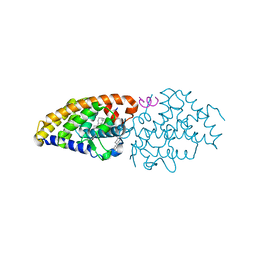 | | Vitamin D Receptor complex with lithocholic acid | | 分子名称: | (3beta,5beta,14beta,17alpha)-3-hydroxycholan-24-oic acid, Nuclear receptor coactivator 2, Vitamin D3 receptor A | | 著者 | Belorusova, A, Rochel, N. | | 登録日 | 2014-04-01 | | 公開日 | 2014-07-02 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structural insights into the molecular mechanism of vitamin d receptor activation by lithocholic Acid involving a new mode of ligand recognition.
J.Med.Chem., 57, 2014
|
|
5M24
 
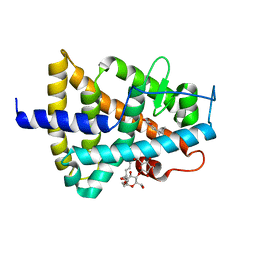 | | RARg mutant-S371E | | 分子名称: | (9cis)-retinoic acid, CHLORIDE ION, DODECYL-ALPHA-D-MALTOSIDE, ... | | 著者 | Rochel, N, Sirigu, S. | | 登録日 | 2016-10-11 | | 公開日 | 2017-09-20 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.69 Å) | | 主引用文献 | Allosteric Regulation in the Ligand Binding Domain of Retinoic Acid Receptor gamma.
PLoS ONE, 12, 2017
|
|
3A9E
 
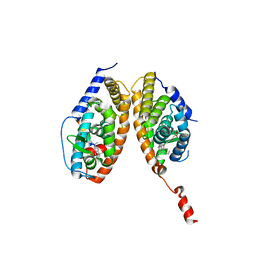 | | Crystal structure of a mixed agonist-bound RAR-alpha and antagonist-bound RXR-alpha heterodimer ligand binding domains | | 分子名称: | (2E,4E,6Z)-3-methyl-7-(5,5,8,8-tetramethyl-3-propoxy-5,6,7,8-tetrahydronaphthalen-2-yl)octa-2,4,6-trienoic acid, 13-mer (LXXLL motif) from Nuclear receptor coactivator 2, RETINOIC ACID, ... | | 著者 | Sato, Y, Duclaud, S, Peluso-Iltis, C, Poussin, P, Moras, D, Rochel, N, Structural Proteomics in Europe (SPINE) | | 登録日 | 2009-10-24 | | 公開日 | 2010-10-06 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.75 Å) | | 主引用文献 | The Phantom Effect of the Rexinoid LG100754: structural and functional insights
Plos One, 5, 2010
|
|
7QPI
 
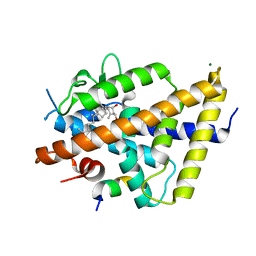 | | Structure of lamprey VDR in complex with 1,25D3 | | 分子名称: | 5-{2-[1-(5-HYDROXY-1,5-DIMETHYL-HEXYL)-7A-METHYL-OCTAHYDRO-INDEN-4-YLIDENE]-ETHYLIDENE}-4-METHYLENE-CYCLOHEXANE-1,3-DIOL, MAGNESIUM ION, Nuclear receptor coactivator 1, ... | | 著者 | Rochel, N. | | 登録日 | 2022-01-04 | | 公開日 | 2022-12-14 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Advances in Vitamin D Receptor Function and Evolution Based on the 3D Structure of the Lamprey Ligand-Binding Domain.
J.Med.Chem., 65, 2022
|
|
7QPP
 
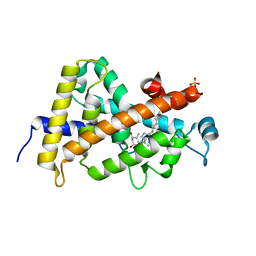 | |
4TR7
 
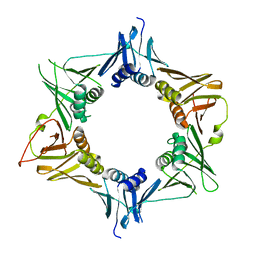 | |
4TR8
 
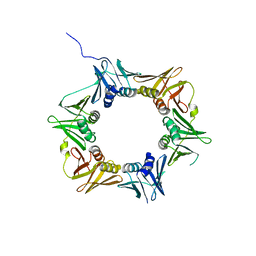 | | Crystal structure of DNA polymerase sliding clamp from Pseudomonas aeruginosa | | 分子名称: | DNA polymerase III subunit beta, SODIUM ION | | 著者 | Olieric, V, Burnouf, D, Ennifar, E, Wolff, P. | | 登録日 | 2014-06-15 | | 公開日 | 2014-09-10 | | 最終更新日 | 2014-12-24 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Differential Modes of Peptide Binding onto Replicative Sliding Clamps from Various Bacterial Origins.
J.Med.Chem., 57, 2014
|
|
4TR6
 
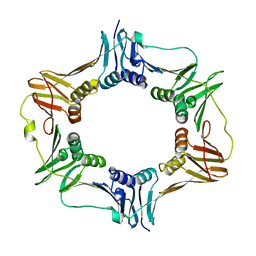 | | Crystal structure of DNA polymerase sliding clamp from Bacillus subtilis | | 分子名称: | DNA polymerase III subunit beta, SODIUM ION | | 著者 | Burnouf, D, Olieric, V, Ennifar, E, Wolff, P. | | 登録日 | 2014-06-14 | | 公開日 | 2014-09-10 | | 最終更新日 | 2014-11-19 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Differential Modes of Peptide Binding onto Replicative Sliding Clamps from Various Bacterial Origins.
J.Med.Chem., 57, 2014
|
|
4TSZ
 
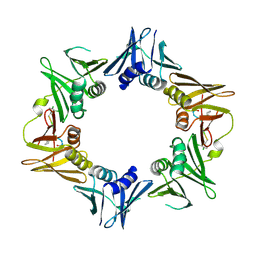 | | Crystal structure of DNA polymerase sliding clamp from Pseudomonas aeruginosa with ligand | | 分子名称: | ACE-GLN-ALC-ASP-LEU-ZCL peptide, DNA polymerase III subunit beta | | 著者 | Olieric, V, Burnouf, D, Ennifar, E, Wolff, P. | | 登録日 | 2014-06-19 | | 公開日 | 2014-09-10 | | 最終更新日 | 2016-12-21 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Differential Modes of Peptide Binding onto Replicative Sliding Clamps from Various Bacterial Origins.
J.Med.Chem., 57, 2014
|
|
6FZJ
 
 | | PPAR gamma mutant complex | | 分子名称: | (2~{S})-3-[4-[2-[methyl(pyridin-2-yl)amino]ethoxy]phenyl]-2-[[2-(phenylcarbonyl)phenyl]amino]propanoic acid, CHLORIDE ION, IODIDE ION, ... | | 著者 | Rochel, N. | | 登録日 | 2018-03-14 | | 公開日 | 2019-02-13 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.011 Å) | | 主引用文献 | Recurrent activating mutations of PPAR gamma associated with luminal bladder tumors.
Nat Commun, 10, 2019
|
|
6FZY
 
 | | PPAR mutant | | 分子名称: | Peroxisome proliferator-activated receptor gamma | | 著者 | Rochel, N. | | 登録日 | 2018-03-15 | | 公開日 | 2019-02-13 | | 実験手法 | X-RAY DIFFRACTION (3.1 Å) | | 主引用文献 | Recurrent activating mutations of PPAR gamma associated with luminal bladder tumors.
Nat Commun, 10, 2019
|
|
6FZG
 
 | | PPAR gamma mutant complex | | 分子名称: | (2~{S})-3-[4-[2-[methyl(pyridin-2-yl)amino]ethoxy]phenyl]-2-[[2-(phenylcarbonyl)phenyl]amino]propanoic acid, CHLORIDE ION, Peroxisome proliferator-activated receptor gamma | | 著者 | Rochel, N. | | 登録日 | 2018-03-14 | | 公開日 | 2019-02-13 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Recurrent activating mutations of PPAR gamma associated with luminal bladder tumors.
Nat Commun, 10, 2019
|
|
6FZF
 
 | | PPAR mutant complex | | 分子名称: | (2~{S})-3-[4-[2-[methyl(pyridin-2-yl)amino]ethoxy]phenyl]-2-[[2-(phenylcarbonyl)phenyl]amino]propanoic acid, Peroxisome proliferator-activated receptor gamma, Peroxisome proliferator-activated receptor gamma coactivator 1-alpha | | 著者 | Rochel, N. | | 登録日 | 2018-03-14 | | 公開日 | 2019-02-13 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Recurrent activating mutations of PPAR gamma associated with luminal bladder tumors.
Nat Commun, 10, 2019
|
|
6FZP
 
 | | PPAR gamma complex. | | 分子名称: | (2~{S})-3-[4-[2-[methyl(pyridin-2-yl)amino]ethoxy]phenyl]-2-[[2-(phenylcarbonyl)phenyl]amino]propanoic acid, Peroxisome proliferator-activated receptor gamma, Peroxisome proliferator-activated receptor gamma coactivator 1-alpha | | 著者 | Rochel, N, Beji, S. | | 登録日 | 2018-03-15 | | 公開日 | 2019-02-13 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Recurrent activating mutations of PPAR gamma associated with luminal bladder tumors.
Nat Commun, 10, 2019
|
|
4G1D
 
 | | Structural basis for the accommodation of bis- and tris-aromatic derivatives in Vitamin D Nuclear Receptor | | 分子名称: | 3-(5'-{[3,4-bis(hydroxymethyl)benzyl]oxy}-2'-methyl-2-propylbiphenyl-4-yl)pentan-3-ol, Nuclear receptor coactivator 1, Vitamin D3 receptor A | | 著者 | Ciesielski, F, Sato, Y, Moras, D, Rochel, N. | | 登録日 | 2012-07-10 | | 公開日 | 2012-09-26 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Structural basis for the accommodation of bis- and tris-aromatic derivatives in vitamin d nuclear receptor.
J.Med.Chem., 55, 2012
|
|
4G20
 
 | | Structural basis for the accommodation of bis- and tris-aromatic derivatives in Vitamin D Nuclear Receptor | | 分子名称: | 3-(5'-{2-[3,4-bis(hydroxymethyl)phenyl]ethyl}-2'-methyl-2-propylbiphenyl-4-yl)pentan-3-ol, Nuclear receptor coactivator 1, Vitamin D3 receptor A | | 著者 | Ciesielski, F, Sato, Y, Moras, D, Rochel, N. | | 登録日 | 2012-07-11 | | 公開日 | 2012-09-26 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Structural basis for the accommodation of bis- and tris-aromatic derivatives in vitamin d nuclear receptor.
J.Med.Chem., 55, 2012
|
|
4G21
 
 | | Structural basis for the accommodation of bis- and tris-aromatic derivatives in Vitamin D Nuclear Receptor | | 分子名称: | 3-(5'-{[3,4-bis(hydroxymethyl)benzyl]oxy}-2'-ethyl-2-propylbiphenyl-4-yl)pentan-3-ol, Nuclear receptor coactivator 1, Vitamin D3 receptor A | | 著者 | Ciesielski, F, Sato, Y, Moras, D, Rochel, N. | | 登録日 | 2012-07-11 | | 公開日 | 2012-09-26 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Structural basis for the accommodation of bis- and tris-aromatic derivatives in vitamin d nuclear receptor.
J.Med.Chem., 55, 2012
|
|
4G2I
 
 | | Structural basis for the accommodation of bis- and tris-aromatic derivatives in Vitamin D Nuclear Receptor | | 分子名称: | (3E,5E)-6-(3-{2-[3,4-bis(hydroxymethyl)phenyl]ethyl}phenyl)-1,1,1-trifluoro-2-(trifluoromethyl)octa-3,5-dien-2-ol, Vitamin D3 receptor | | 著者 | Ciesielski, F, Sato, Y, Moras, D, Rochel, N. | | 登録日 | 2012-07-12 | | 公開日 | 2012-09-26 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structural basis for the accommodation of bis- and tris-aromatic derivatives in vitamin d nuclear receptor.
J.Med.Chem., 55, 2012
|
|
