6GHS
 
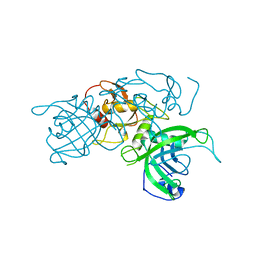 | | Modification dependent TagI restriction endonuclease | | Descriptor: | SODIUM ION, TagI restriction endonuclease, ZINC ION | | Authors: | Kisiala, M, Copelas, A, Czapinska, H, Xu, S, Bochtler, M. | | Deposit date: | 2018-05-08 | | Release date: | 2018-08-29 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.92 Å) | | Cite: | Crystal structure of the modification-dependent SRA-HNH endonuclease TagI.
Nucleic Acids Res., 46, 2018
|
|
3FC3
 
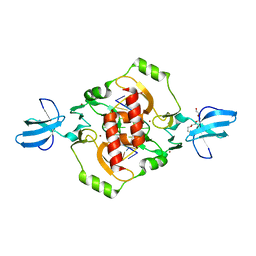 | | Crystal structure of the beta-beta-alpha-Me type II restriction endonuclease Hpy99I | | Descriptor: | 5'-(*DCP*DTP*DCP*DGP*DAP*DCP*DGP*DTP*DAP*DGP*DA)-3', 5'-(*DTP*DAP*DCP*DGP*DTP*DCP*DGP*DAP*DGP*DTP*DC)-3', Restriction endonuclease Hpy99I, ... | | Authors: | Sokolowska, M, Czapinska, H, Bochtler, M. | | Deposit date: | 2008-11-21 | | Release date: | 2009-03-31 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of the beta beta alpha-Me type II restriction endonuclease Hpy99I with target DNA.
Nucleic Acids Res., 37, 2009
|
|
4KYW
 
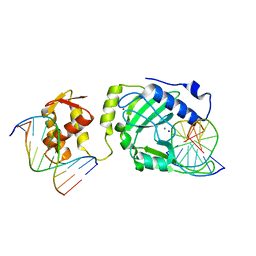 | | Restriction endonuclease DPNI in complex with two DNA molecules | | Descriptor: | 5'-(*DC*DTP*DGP*DGP*6MAP*DTP*DCP*DCP*DAP*DG)-3', CALCIUM ION, SODIUM ION, ... | | Authors: | Mierzejewska, K, Siwek, W, Czapinska, H, Skowronek, K, Bujnicki, J.M, Bochtler, M. | | Deposit date: | 2013-05-29 | | Release date: | 2014-06-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural basis of the methylation specificity of R.DpnI.
Nucleic Acids Res., 42, 2014
|
|
4LXC
 
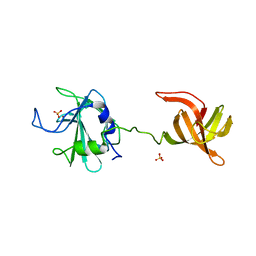 | | The antimicrobial peptidase lysostaphin from Staphylococcus simulans | | Descriptor: | Lysostaphin, SULFATE ION, ZINC ION | | Authors: | Sabala, I, Jagielska, E, Bardelang, P.T, Czapinska, H, Dahms, S.O, Sharpe, J.A, James, R, Than, M.E, Thomas, N.R, Bochtler, M. | | Deposit date: | 2013-07-29 | | Release date: | 2014-07-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Crystal structure of the antimicrobial peptidase lysostaphin from Staphylococcus simulans.
Febs J., 281, 2014
|
|
4OQ2
 
 | | 5hmC specific restriction endonuclease PvuRTs1I | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Restriction endonuclease PvuRts1 I | | Authors: | Kazrani, A.A, Kowalska, M, Czapinska, H, Bochtler, M. | | Deposit date: | 2014-02-07 | | Release date: | 2014-03-12 | | Last modified: | 2018-03-07 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal structure of the 5hmC specific endonuclease PvuRts1I.
Nucleic Acids Res., 42, 2014
|
|
4QP5
 
 | | Catalytic domain of the antimicrobial peptidase lysostaphin from Staphylococcus simulans crystallized in the presence of phosphate | | Descriptor: | GLYCEROL, Lysostaphin, PHOSPHATE ION, ... | | Authors: | Sabala, I, Jagielska, E, Bardelang, P.T, Czapinska, H, Dahms, S.O, Sharpe, J.A, James, R, Than, M.E, Thomas, N.R, Bochtler, M. | | Deposit date: | 2014-06-22 | | Release date: | 2014-07-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.26 Å) | | Cite: | Crystal structure of the antimicrobial peptidase lysostaphin from Staphylococcus simulans.
Febs J., 281, 2014
|
|
6S58
 
 | | AvaII restriction endonuclease in the absence of nucleic acids | | Descriptor: | CALCIUM ION, Type II site-specific deoxyribonuclease, UNKNOWN ATOM OR ION | | Authors: | Kisiala, M, Kowalska, M, Korza, H, Czapinska, H, Bochtler, M. | | Deposit date: | 2019-06-30 | | Release date: | 2020-05-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Restriction endonucleases that cleave RNA/DNA heteroduplexes bind dsDNA in A-like conformation.
Nucleic Acids Res., 48, 2020
|
|
6S48
 
 | | AvaII RESTRICTION ENDONUCLEASE IN COMPLEX WITH PARTIALLY CLEAVED dsDNA | | Descriptor: | BETA-MERCAPTOETHANOL, CALCIUM ION, DNA (5'-D(*GP*AP*TP*G)-3'), ... | | Authors: | Kisiala, M, Kowalska, M, Korza, H, Czapinska, H, Bochtler, M. | | Deposit date: | 2019-06-26 | | Release date: | 2020-05-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Restriction endonucleases that cleave RNA/DNA heteroduplexes bind dsDNA in A-like conformation.
Nucleic Acids Res., 48, 2020
|
|
3NDH
 
 | | Restriction endonuclease in complex with substrate DNA | | Descriptor: | ACETATE ION, CHLORIDE ION, DNA (5'-D(*C*CP*AP*TP*CP*GP*CP*GP*TP*AP*C)-3'), ... | | Authors: | Firczuk, M, Wojciechowski, M, Czapinska, H, Bochtler, M. | | Deposit date: | 2010-06-07 | | Release date: | 2010-07-28 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | DNA intercalation without flipping in the specific ThaI-DNA complex
Nucleic Acids Res., 39, 2011
|
|
3OQG
 
 | | Restriction endonuclease HPY188I in complex with substrate DNA | | Descriptor: | CHLORIDE ION, DNA 5'-D(*GP*AP*TP*CP*TP*GP*AP*AP*C)-3', DNA 5'-D(*GP*TP*TP*CP*AP*GP*AP*TP*C)-3', ... | | Authors: | Sokolowska, M, Czapinska, H, Bochtler, M. | | Deposit date: | 2010-09-03 | | Release date: | 2010-10-20 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Hpy188I-DNA pre- and post-cleavage complexes--snapshots of the GIY-YIG nuclease mediated catalysis.
Nucleic Acids Res., 39, 2011
|
|
3PRH
 
 | | tryptophanyl-tRNA synthetase Val144Pro mutant from B. subtilis | | Descriptor: | Tryptophanyl-tRNA synthetase | | Authors: | Antonczak, A.K, Simova, Z, Yonemoto, I, Bochtler, M, Piasecka, A, Czapinska, H, Brancale, A, Tippmann, E.M. | | Deposit date: | 2010-11-29 | | Release date: | 2011-01-19 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Importance of single molecular determinants in the fidelity of expanded genetic codes.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
6R64
 
 | | N-terminal domain of modification dependent EcoKMcrA restriction endonuclease (NEco) in complex with C5mCGG target sequence | | Descriptor: | 5-methylcytosine-specific restriction enzyme A, DNA (5'-D(*GP*AP*AP*CP*(5CM)P*GP*GP*TP*GP*A)-3'), DNA (5'-D(*TP*CP*AP*CP*(5CM)P*GP*GP*TP*TP*C)-3') | | Authors: | Slyvka, A, Zagorskaite, E, Czapinska, H, Sasnauskas, G, Bochtler, M. | | Deposit date: | 2019-03-26 | | Release date: | 2019-10-23 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Crystal structure of the EcoKMcrA N-terminal domain (NEco): recognition of modified cytosine bases without flipping.
Nucleic Acids Res., 47, 2019
|
|
6T22
 
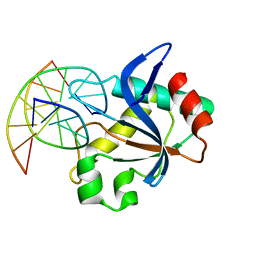 | | N-terminal domain of EcoKMcrA restriction endonuclease (NEco) in complex with T5hmCGA target sequence | | Descriptor: | DNA (5'-D(*GP*AP*AP*TP*(5HC)P*GP*AP*TP*GP*A)-3'), DNA (5'-D(*TP*CP*AP*TP*(5HC)P*GP*AP*TP*TP*C)-3'), EcoKMcrA modification dependent restriction endonuclease | | Authors: | Slyvka, A, Zagorskaite, E, Czapinska, H, Sasnauskas, G, Bochtler, M. | | Deposit date: | 2019-10-07 | | Release date: | 2019-10-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Crystal structure of the EcoKMcrA N-terminal domain (NEco): recognition of modified cytosine bases without flipping.
Nucleic Acids Res., 47, 2019
|
|
6T21
 
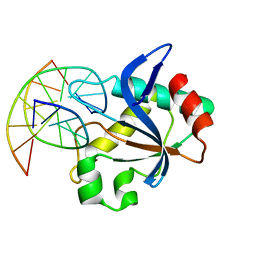 | | N-terminal domain of EcoKMcrA restriction endonuclease (NEco) in complex with T5mCGA target sequence | | Descriptor: | 5-methylcytosine-specific restriction enzyme A, DNA (5'-D(*GP*AP*AP*TP*(5CM)P*GP*AP*TP*GP*A)-3'), DNA (5'-D(*TP*CP*AP*TP*(5CM)P*GP*AP*TP*TP*C)-3') | | Authors: | Slyvka, A, Zagorskaite, E, Czapinska, H, Sasnauskas, G, Bochtler, M. | | Deposit date: | 2019-10-07 | | Release date: | 2019-10-23 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Crystal structure of the EcoKMcrA N-terminal domain (NEco): recognition of modified cytosine bases without flipping.
Nucleic Acids Res., 47, 2019
|
|
1P2N
 
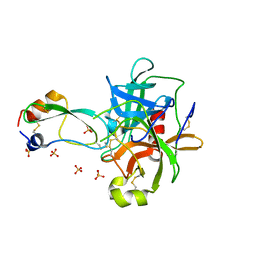 | | Structural consequences of accommodation of four non-cognate amino-acid residues in the S1 pocket of bovine trypsin and chymotrypsin | | Descriptor: | Chymotrypsinogen A, Pancreatic trypsin inhibitor, SULFATE ION | | Authors: | Helland, R, Czapinska, H, Leiros, I, Olufsen, M, Otlewski, J, Smalaas, A.O. | | Deposit date: | 2003-04-15 | | Release date: | 2004-04-20 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural consequences of accommodation of four non-cognate amino acid residues in the S1 pocket of bovine trypsin and chymotrypsin.
J.Mol.Biol., 333, 2003
|
|
1P2I
 
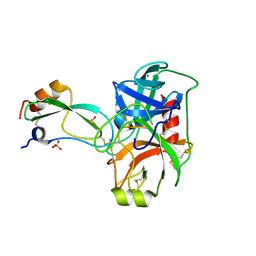 | | Structural consequences of accommodation of four non-cognate amino-acid residues in the S1 pocket of bovine trypsin and chymotrypsin | | Descriptor: | CALCIUM ION, Pancreatic trypsin inhibitor, SULFATE ION, ... | | Authors: | Helland, R, Czapinska, H, Leiros, I, Olufsen, M, Otlewski, J, Smalaas, A.O. | | Deposit date: | 2003-04-15 | | Release date: | 2004-04-20 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural consequences of accommodation of four non-cognate amino acid residues in the S1 pocket of bovine trypsin and chymotrypsin.
J.Mol.Biol., 333, 2003
|
|
1P2O
 
 | | Structural consequences of accommodation of four non-cognate amino-acid residues in the S1 pocket of bovine trypsin and chymotrypsin | | Descriptor: | Chymotrypsinogen A, Pancreatic trypsin inhibitor, SULFATE ION | | Authors: | Helland, R, Czapinska, H, Leiros, I, Olufsen, M, Otlewski, J, Smalaas, A.O. | | Deposit date: | 2003-04-15 | | Release date: | 2004-04-20 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural consequences of accommodation of four non-cognate amino acid residues in the S1 pocket of bovine trypsin and chymotrypsin.
J.Mol.Biol., 333, 2003
|
|
1P2M
 
 | | Structural consequences of accommodation of four non-cognate amino-acid residues in the S1 pocket of bovine trypsin and chymotrypsin | | Descriptor: | Chymotrypsinogen A, Pancreatic trypsin inhibitor, SULFATE ION | | Authors: | Helland, R, Czapinska, H, Leiros, I, Olufsen, M, Otlewski, J, Smalaas, A.O. | | Deposit date: | 2003-04-15 | | Release date: | 2004-04-20 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural consequences of accommodation of four non-cognate amino acid residues in the S1 pocket of bovine trypsin and chymotrypsin.
J.Mol.Biol., 333, 2003
|
|
1P2J
 
 | | Structural consequences of accommodation of four non-cognate amino-acid residues in the S1 pocket of bovine trypsin and chymotrypsin | | Descriptor: | CALCIUM ION, Pancreatic trypsin inhibitor, SULFATE ION, ... | | Authors: | Helland, R, Czapinska, H, Leiros, I, Olufsen, M, Otlewski, J, Smalaas, A.O. | | Deposit date: | 2003-04-15 | | Release date: | 2004-04-20 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structural consequences of accommodation of four non-cognate amino acid residues in the S1 pocket of bovine trypsin and chymotrypsin.
J.Mol.Biol., 333, 2003
|
|
1P2K
 
 | | Structural consequences of accommodation of four non-cognate amino-acid residues in the S1 pocket of bovine trypsin and chymotrypsin | | Descriptor: | CALCIUM ION, Pancreatic trypsin inhibitor, SULFATE ION, ... | | Authors: | Helland, R, Czapinska, H, Leiros, I, Olufsen, M, Otlewski, J, Smalaas, A.O. | | Deposit date: | 2003-04-15 | | Release date: | 2004-04-20 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural consequences of accommodation of four non-cognate amino acid residues in the S1 pocket of bovine trypsin and chymotrypsin.
J.Mol.Biol., 333, 2003
|
|
1P2Q
 
 | | Structural consequences of accommodation of four non-cognate amino-acid residues in the S1 pocket of bovine trypsin and chymotrypsin | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Chymotrypsinogen A, Pancreatic trypsin inhibitor, ... | | Authors: | Helland, R, Czapinska, H, Leiros, I, Olufsen, M, Otlewski, J, Smalaas, A.O. | | Deposit date: | 2003-04-15 | | Release date: | 2004-04-20 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural consequences of accommodation of four non-cognate amino acid residues in the S1 pocket of bovine trypsin and chymotrypsin.
J.Mol.Biol., 333, 2003
|
|
3FUC
 
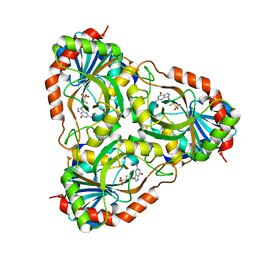 | | Recombinant calf purine nucleoside phosphorylase in a binary complex with multisubstrate analogue inhibitor 9-(5',5'-difluoro-5'-phosphonopentyl)-9-deazaguanine structure in a new space group with one full trimer in the asymmetric unit | | Descriptor: | AZIDE ION, MAGNESIUM ION, Purine nucleoside phosphorylase, ... | | Authors: | Bochtler, M, Breer, K, Bzowska, A, Chojnowski, G, Hashimoto, M, Hikishima, S, Narczyk, M, Wielgus-Kutrowska, B, Yokomatsu, T. | | Deposit date: | 2009-01-14 | | Release date: | 2009-12-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | 1.45 A resolution crystal structure of recombinant PNP in complex with a pM multisubstrate analogue inhibitor bearing one feature of the postulated transition state.
Biochem.Biophys.Res.Commun., 391, 2010
|
|
8UOU
 
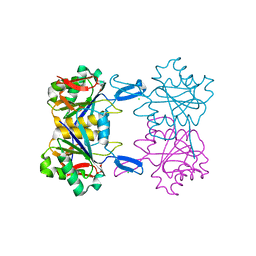 | | Structure of atypical asparaginase from Rhodospirillum rubrum in complex with L-Asp | | Descriptor: | ASPARTIC ACID, Asparaginase, CHLORIDE ION | | Authors: | Lubkowski, J, Wlodawer, A, Zhang, D. | | Deposit date: | 2023-10-20 | | Release date: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | RrA, an enzyme from Rhodospirillum rubrum, is a prototype of a new family of short-chain L-asparaginases.
Protein Sci., 33, 2024
|
|
8UPC
 
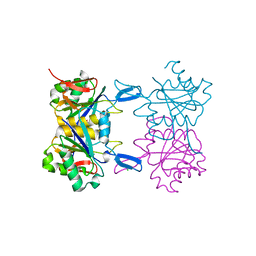 | | Structure of atypical asparaginase from Rhodospirillum rubrum (mutant K158M) | | Descriptor: | Asparaginase, CHLORIDE ION, GLYCEROL | | Authors: | Lubkowski, J, Wlodawer, A, Zhang, D. | | Deposit date: | 2023-10-22 | | Release date: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | RrA, an enzyme from Rhodospirillum rubrum, is a prototype of a new family of short-chain L-asparaginases.
Protein Sci., 33, 2024
|
|
8UP3
 
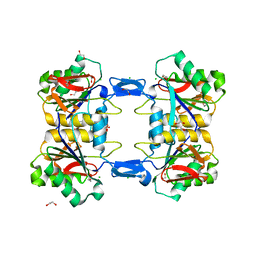 | | Structure of atypical asparaginase from Rhodospirillum rubrum (mutant Y21F) | | Descriptor: | 1,2-ETHANEDIOL, ASPARTIC ACID, Asparaginase, ... | | Authors: | Lubkowski, J, Wlodawer, A, Zhang, D. | | Deposit date: | 2023-10-20 | | Release date: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | RrA, an enzyme from Rhodospirillum rubrum, is a prototype of a new family of short-chain L-asparaginases.
Protein Sci., 33, 2024
|
|
