5KTB
 
 | |
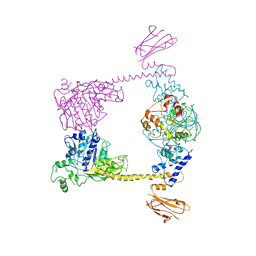
2Q2E
 
 | |
1MU5
 
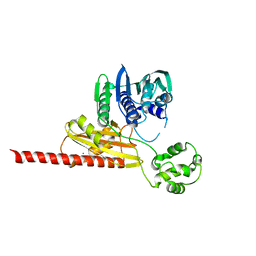 | | Structure of topoisomerase subunit | | Descriptor: | CALCIUM ION, Type II DNA topoisomerase VI Subunit B | | Authors: | Corbett, K.D, Berger, J.M. | | Deposit date: | 2002-09-23 | | Release date: | 2003-01-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the topoisomerase VI-B subunit: implications for type II topoisomerase mechanism and evolution
Embo J., 22, 2003
|
|
1MX0
 
 | | Structure of topoisomerase subunit | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, SODIUM ION, ... | | Authors: | Corbett, K.D, Berger, J.M. | | Deposit date: | 2002-10-01 | | Release date: | 2003-01-07 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of the topoisomerase VI-B subunit: implications for type II topoisomerase mechanism and evolution
Embo J., 22, 2003
|
|
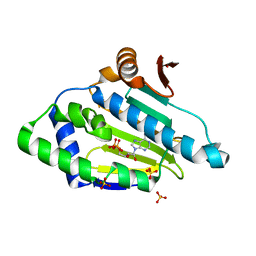
3K60
 
 | |
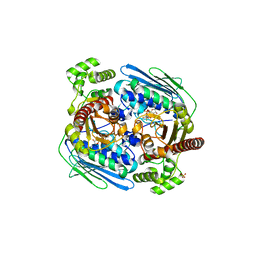
1Z5B
 
 | |
1Z59
 
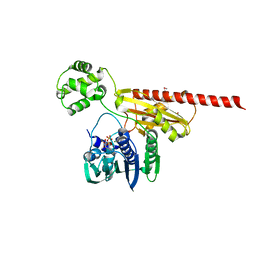 | | Topoisomerase VI-B, ADP-bound monomer form | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Type II DNA topoisomerase VI subunit B | | Authors: | Corbett, K.D, Berger, J.M. | | Deposit date: | 2005-03-17 | | Release date: | 2005-06-14 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural dissection of ATP turnover in the prototypical GHL ATPase TopoVI.
Structure, 13, 2005
|
|
1Z5A
 
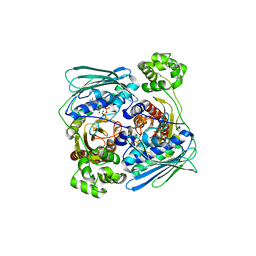 | | Topoisomerase VI-B, ADP-bound dimer form | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Type II DNA topoisomerase VI subunit B | | Authors: | Corbett, K.D, Berger, J.M. | | Deposit date: | 2005-03-17 | | Release date: | 2005-06-14 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural dissection of ATP turnover in the prototypical GHL ATPase TopoVI.
Structure, 13, 2005
|
|
1Z5C
 
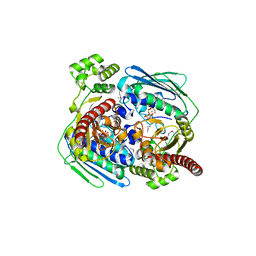 | | Topoisomerase VI-B, ADP Pi bound dimer form | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Corbett, K.D, Berger, J.M. | | Deposit date: | 2005-03-17 | | Release date: | 2005-06-14 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural dissection of ATP turnover in the prototypical GHL ATPase TopoVI.
Structure, 13, 2005
|
|
1ZVU
 
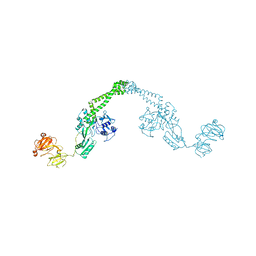 | |
1ZVT
 
 | |
2HKJ
 
 | | Topoisomerase VI-B bound to radicicol | | Descriptor: | DIMETHYL SULFOXIDE, MAGNESIUM ION, RADICICOL, ... | | Authors: | Corbett, K.D, Berger, J.M. | | Deposit date: | 2006-07-04 | | Release date: | 2006-08-29 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for topoisomerase VI inhibition
by the anti-Hsp90 drug radicicol
Nucleic Acids Res., 34, 2006
|
|
3N4S
 
 | | Structure of Csm1 C-terminal domain, P21212 form | | Descriptor: | Monopolin complex subunit CSM1, PENTAETHYLENE GLYCOL | | Authors: | Corbett, K.D, Harrison, S.C. | | Deposit date: | 2010-05-22 | | Release date: | 2010-09-01 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | The Monopolin Complex Crosslinks Kinetochore Components to Regulate Chromosome-Microtubule Attachments.
Cell(Cambridge,Mass.), 142, 2010
|
|
3N7N
 
 | | Structure of Csm1/Lrs4 complex | | Descriptor: | Monopolin complex subunit CSM1, Monopolin complex subunit LRS4 | | Authors: | Corbett, K.D, Harrison, S.C. | | Deposit date: | 2010-05-27 | | Release date: | 2010-09-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.9 Å) | | Cite: | The Monopolin Complex Crosslinks Kinetochore Components to Regulate Chromosome-Microtubule Attachments.
Cell(Cambridge,Mass.), 142, 2010
|
|
3N4R
 
 | | Structure of Csm1 C-terminal domain, R3 form | | Descriptor: | MALONATE ION, Monopolin complex subunit CSM1, PENTAETHYLENE GLYCOL | | Authors: | Corbett, K.D, Harrison, S.C. | | Deposit date: | 2010-05-22 | | Release date: | 2010-09-01 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.602 Å) | | Cite: | The Monopolin Complex Crosslinks Kinetochore Components to Regulate Chromosome-Microtubule Attachments.
Cell(Cambridge,Mass.), 142, 2010
|
|
3N4X
 
 | | Structure of Csm1 full-length | | Descriptor: | Monopolin complex subunit CSM1 | | Authors: | Corbett, K.D, Harrison, S.C. | | Deposit date: | 2010-05-23 | | Release date: | 2010-09-01 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.408 Å) | | Cite: | The Monopolin Complex Crosslinks Kinetochore Components to Regulate Chromosome-Microtubule Attachments.
Cell(Cambridge,Mass.), 142, 2010
|
|
1SUU
 
 | |
7R98
 
 | |
6U7B
 
 | | Structure of E. coli MS115-1 CdnC:HORMA-deltaN complex | | Descriptor: | ACETATE ION, ADENOSINE MONOPHOSPHATE, CHLORIDE ION, ... | | Authors: | Ye, Q, Corbett, K.D. | | Deposit date: | 2019-09-02 | | Release date: | 2019-12-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | HORMA Domain Proteins and a Trip13-like ATPase Regulate Bacterial cGAS-like Enzymes to Mediate Bacteriophage Immunity.
Mol.Cell, 77, 2020
|
|
6BZG
 
 | | Structure of S. cerevisiae Zip2:Spo16 complex, P212121 form | | Descriptor: | HEXAETHYLENE GLYCOL, Protein ZIP2, SULFATE ION, ... | | Authors: | Arora, K, Corbett, K.D. | | Deposit date: | 2017-12-23 | | Release date: | 2018-02-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | The conserved XPF:ERCC1-like Zip2:Spo16 complex controls meiotic crossover formation through structure-specific DNA binding.
Nucleic Acids Res., 47, 2019
|
|
6BZF
 
 | | Structure of S. cerevisiae Zip2:Spo16 complex, C2 form | | Descriptor: | Protein ZIP2, Sporulation-specific protein 16 | | Authors: | Arora, K, Corbett, K.D. | | Deposit date: | 2017-12-23 | | Release date: | 2018-02-14 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.286 Å) | | Cite: | The conserved XPF:ERCC1-like Zip2:Spo16 complex controls meiotic crossover formation through structure-specific DNA binding.
Nucleic Acids Res., 47, 2019
|
|
4IM8
 
 | | low resolution crystal structure of mouse RAGE | | Descriptor: | Advanced glycation end-products receptor | | Authors: | Xu, D, Young, J.H, Krahn, J.M, Song, D, Corbett, K.D, Chazin, W.J, Pedersen, L.C, Esko, J.D. | | Deposit date: | 2013-01-02 | | Release date: | 2013-08-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.503 Å) | | Cite: | Stable RAGE-Heparan Sulfate Complexes Are Essential for Signal Transduction.
Acs Chem.Biol., 8, 2013
|
|
6WZO
 
 | |
6WZQ
 
 | |
6MJC
 
 | |
