7WKA
 
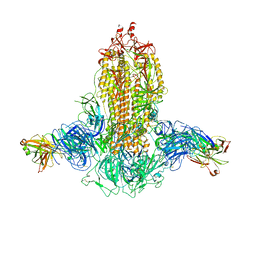 | |
7WEV
 
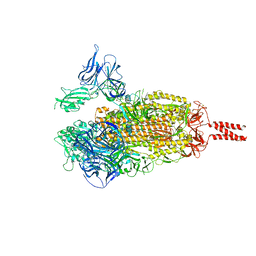 | |
7WK9
 
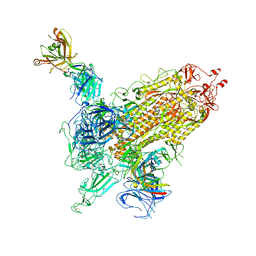 | |
7WVO
 
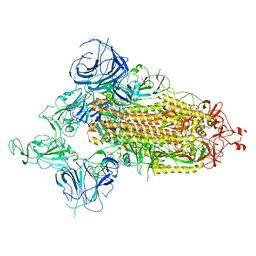 | | SARS-CoV-2 Omicron S-open-2 | | 分子名称: | Spike glycoprotein | | 著者 | Li, J.W, Cong, Y. | | 登録日 | 2022-02-10 | | 公開日 | 2022-03-02 | | 最終更新日 | 2022-05-04 | | 実験手法 | ELECTRON MICROSCOPY (3.41 Å) | | 主引用文献 | Molecular basis of receptor binding and antibody neutralization of Omicron.
Nature, 604, 2022
|
|
7WVN
 
 | | SARS-CoV-2 Omicron S-open | | 分子名称: | Spike glycoprotein | | 著者 | Li, J.W, Cong, Y. | | 登録日 | 2022-02-10 | | 公開日 | 2022-03-02 | | 最終更新日 | 2022-05-04 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Molecular basis of receptor binding and antibody neutralization of Omicron.
Nature, 604, 2022
|
|
7WK3
 
 | | SARS-CoV-2 Omicron S-open | | 分子名称: | Spike glycoprotein | | 著者 | Li, J.W, Cong, Y. | | 登録日 | 2022-01-08 | | 公開日 | 2022-01-26 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Structural basis for ACE2 engagement and antibody evasion and neutralization of SARS-Co-2 Omicron varient
To Be Published
|
|
7V5G
 
 | | 20S+monoUb-CyclinB1-NT (S1) | | 分子名称: | Proteasome subunit alpha type-1, Proteasome subunit alpha type-2, Proteasome subunit alpha type-3, ... | | 著者 | Xu, C, Cong, Y. | | 登録日 | 2021-08-17 | | 公開日 | 2021-09-22 | | 最終更新日 | 2022-03-30 | | 実験手法 | ELECTRON MICROSCOPY (4.47 Å) | | 主引用文献 | The 20S as a stand-alone proteasome in cells can degrade the ubiquitin tag.
Nat Commun, 12, 2021
|
|
7V5M
 
 | | 20S+monoUb-CyclinB1-NT (S2) | | 分子名称: | Proteasome subunit alpha type-1, Proteasome subunit alpha type-2, Proteasome subunit alpha type-3, ... | | 著者 | Xu, C, Cong, Y. | | 登録日 | 2021-08-17 | | 公開日 | 2021-09-22 | | 最終更新日 | 2022-03-30 | | 実験手法 | ELECTRON MICROSCOPY (3.88 Å) | | 主引用文献 | The 20S as a stand-alone proteasome in cells can degrade the ubiquitin tag.
Nat Commun, 12, 2021
|
|
7YS6
 
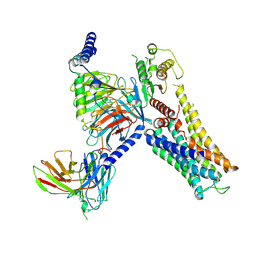 | | Cryo-EM structure of the Serotonin 6 (5-HT6) receptor-DNGs-scFv16 complex | | 分子名称: | 5-hydroxytryptamine receptor 6, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | 著者 | Zhao, Q.Y, Wang, Y.F, He, L, Wang, S, Cong, Y. | | 登録日 | 2022-08-11 | | 公開日 | 2023-03-29 | | 最終更新日 | 2023-10-11 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | Structural insights into constitutive activity of 5-HT 6 receptor.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
4U7P
 
 | | Crystal structure of DNMT3A-DNMT3L complex | | 分子名称: | DNA (cytosine-5)-methyltransferase 3-like, DNA (cytosine-5)-methyltransferase 3A, S-ADENOSYL-L-HOMOCYSTEINE, ... | | 著者 | Wang, L, Guo, X, Li, J, Xiao, J, Yin, X, He, S, Wang, J, Xu, Y. | | 登録日 | 2014-07-31 | | 公開日 | 2014-11-12 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (3.821 Å) | | 主引用文献 | Structural insight into autoinhibition and histone H3-induced activation of DNMT3A
Nature, 517, 2015
|
|
4U7T
 
 | | Crystal structure of DNMT3A-DNMT3L in complex with histone H3 | | 分子名称: | DNA (cytosine-5)-methyltransferase 3-like, DNA (cytosine-5)-methyltransferase 3A, S-ADENOSYL-L-HOMOCYSTEINE, ... | | 著者 | Guo, X, Wang, L, Yin, X, Li, J, Xiao, J, He, S, Wang, J, Xu, Y. | | 登録日 | 2014-07-31 | | 公開日 | 2014-11-12 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Structural insight into autoinhibition and histone H3-induced activation of DNMT3A
Nature, 517, 2015
|
|
7LQ7
 
 | | Crystal structure of SARS-CoV-2 receptor binding domain in complex with antibodies CV503 and COVA1-16 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, COVA1-16 heavy chain, COVA1-16 light chain, ... | | 著者 | Yuan, M, Zhu, X, Wilson, I.A. | | 登録日 | 2021-02-13 | | 公開日 | 2021-09-15 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (3.4 Å) | | 主引用文献 | Bispecific antibodies targeting distinct regions of the spike protein potently neutralize SARS-CoV-2 variants of concern.
Sci Transl Med, 13, 2021
|
|
2OBD
 
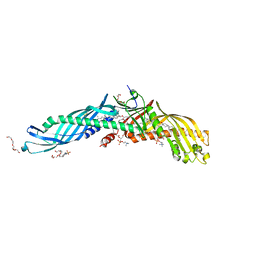 | | Crystal Structure of Cholesteryl Ester Transfer Protein | | 分子名称: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | 著者 | Qiu, X. | | 登録日 | 2006-12-18 | | 公開日 | 2007-01-23 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Crystal structure of cholesteryl ester transfer protein reveals a long tunnel and four bound lipid molecules.
Nat.Struct.Mol.Biol., 14, 2007
|
|
8DAO
 
 | | Crystal structure of SARS-CoV-2 spike stem fusion peptide in complex with neutralizing antibody COV44-79 | | 分子名称: | COV44-79 heavy chain constant domain, COV44-79 heavy chain variable domain, COV44-79 light chain constant domain, ... | | 著者 | Lin, T.H, Lee, C.C.D, Yuan, M, Wilson, I.A. | | 登録日 | 2022-06-13 | | 公開日 | 2022-07-27 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Broadly neutralizing antibodies target the coronavirus fusion peptide.
Science, 377, 2022
|
|
8D6Z
 
 | | Crystal structure of SARS-CoV-2 spike stem fusion peptide in complex with neutralizing antibody COV91-27 | | 分子名称: | Neutralizing antibody COV91-27 heavy chain, Neutralizing antibody COV91-27 light chain, Spike protein S2 fusion peptide | | 著者 | Lee, C.C.D, Lin, T.H, Yuan, M, Wilson, I.A. | | 登録日 | 2022-06-06 | | 公開日 | 2022-07-27 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Broadly neutralizing antibodies target the coronavirus fusion peptide.
Science, 377, 2022
|
|
8D36
 
 | |
8DTT
 
 | | Crystal structure of SARS-CoV-2 spike stem helix peptide in complex with neutralizing antibody COV93-03 | | 分子名称: | COV93-03 heavy chain, COV93-03 light chain, Spike protein S2' stem helix peptide | | 著者 | Lee, C.C.D, Lin, T.H, Wilson, I.A. | | 登録日 | 2022-07-26 | | 公開日 | 2022-11-23 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Rare, convergent antibodies targeting the stem helix broadly neutralize diverse betacoronaviruses.
Cell Host Microbe, 31, 2023
|
|
8DTX
 
 | | Crystal structure of SARS-CoV-2 spike stem helix peptide in complex with neutralizing antibody COV89-22 | | 分子名称: | COV89-22 heavy chain, COV89-22 light chain, Spike protein S2' stem helix peptide | | 著者 | Lin, T.H, Lee, C.C.D, Wilson, I.A. | | 登録日 | 2022-07-26 | | 公開日 | 2022-11-23 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Rare, convergent antibodies targeting the stem helix broadly neutralize diverse betacoronaviruses.
Cell Host Microbe, 31, 2023
|
|
8DTR
 
 | | Crystal structure of SARS-CoV-2 spike stem helix peptide in complex with neutralizing antibody COV30-14 | | 分子名称: | COV30-14 heavy chain, COV30-14 light chain, Spike protein S2' stem helix peptide | | 著者 | Lee, C.C.D, Lin, T.H, Wilson, I.A. | | 登録日 | 2022-07-26 | | 公開日 | 2022-11-23 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Rare, convergent antibodies targeting the stem helix broadly neutralize diverse betacoronaviruses.
Cell Host Microbe, 31, 2023
|
|
7C8V
 
 | | Structure of sybody SR4 in complex with the SARS-CoV-2 S Receptor Binding domain (RBD) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, Spike protein S1, ... | | 著者 | Li, T, Yao, H, Cai, H, Qin, W, Li, D. | | 登録日 | 2020-06-03 | | 公開日 | 2020-06-24 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.15 Å) | | 主引用文献 | A synthetic nanobody targeting RBD protects hamsters from SARS-CoV-2 infection.
Nat Commun, 12, 2021
|
|
7C8W
 
 | | Structure of sybody MR17 in complex with the SARS-CoV-2 S receptor-binding domain (RBD) | | 分子名称: | GLYCEROL, Spike protein S1, Synthetic nanobody MR17, ... | | 著者 | Li, T, Cai, H, Yao, H, Qin, W, Li, D. | | 登録日 | 2020-06-03 | | 公開日 | 2020-06-24 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.77 Å) | | 主引用文献 | A synthetic nanobody targeting RBD protects hamsters from SARS-CoV-2 infection.
Nat Commun, 12, 2021
|
|
7CAN
 
 | | Structure of sybody MR17-K99Y in complex with the SARS-CoV-2 S Receptor-binding domain (RBD) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, Spike protein S1, ... | | 著者 | Li, T, Yao, H, Cai, H, Qin, W, Li, D. | | 登録日 | 2020-06-09 | | 公開日 | 2020-06-24 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.94 Å) | | 主引用文献 | A synthetic nanobody targeting RBD protects hamsters from SARS-CoV-2 infection.
Nat Commun, 12, 2021
|
|
5WBP
 
 | |
5WBM
 
 | |
5WBR
 
 | | Structure of human Ketohexokinase complexed with hits from fragment screening | | 分子名称: | 6-[4-(2-hydroxyethyl)piperazin-1-yl]-2-[(3S)-3-(hydroxymethyl)piperidin-1-yl]-4-(trifluoromethyl)pyridine-3-carbonitrile, CITRIC ACID, GLYCEROL, ... | | 著者 | Pandit, J. | | 登録日 | 2017-06-29 | | 公開日 | 2017-09-13 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.58 Å) | | 主引用文献 | Discovery of Fragment-Derived Small Molecules for in Vivo Inhibition of Ketohexokinase (KHK).
J. Med. Chem., 60, 2017
|
|
