1GNX
 
 | | b-glucosidase from Streptomyces sp | | Descriptor: | BETA-GLUCOSIDASE, SULFATE ION, beta-D-fructofuranose-(2-1)-alpha-D-glucopyranose | | Authors: | Guasch, A, Perez-Pons, J.A, Vallmitjana, M, Querol, E, Coll, M. | | Deposit date: | 2001-10-10 | | Release date: | 2002-10-17 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Beta-Glucosidase from Streptomyces
To be Published
|
|
1S6M
 
 | | Conjugative Relaxase Trwc In Complex With Orit DNA. Metal-Bound Structure | | Descriptor: | DNA (25-MER), NICKEL (II) ION, TrwC | | Authors: | Guasch, A, Lucas, M, Moncalian, G, Cabezas, M, Perez-Luque, R, Gomis-Ruth, F.X, de la Cruz, F, Coll, M. | | Deposit date: | 2004-01-26 | | Release date: | 2005-06-14 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Unveiling the molecular mechanism of a conjugative relaxase: The structure of TrwC complexed with a 27-mer DNA comprising the recognition hairpin and the cleavage site.
J.Mol.Biol., 358, 2006
|
|
2ET0
 
 | | The structure of a three-way DNA junction in complex with a metallo-supramolecular helicate reveals a new target for drugs | | Descriptor: | 5'-D(*CP*GP*TP*AP*CP*G)-3', FE (II) ION, N-[(1E)-PYRIDIN-2-YLMETHYLENE]-N-[4-(4-{[(1E)-PYRIDIN-2-YLMETHYLENE]AMINO}BENZYL)PHENYL]AMINE | | Authors: | Oleksi, A, Blanco, A.G, Boer, R, Uson, I, Aymami, J, Coll, M. | | Deposit date: | 2005-10-27 | | Release date: | 2006-03-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Molecular Recognition of a Three-Way DNA Junction by a Metallosupramolecular Helicate
ANGEW.CHEM.INT.ED.ENGL., 45, 2006
|
|
2GLR
 
 | |
1PCA
 
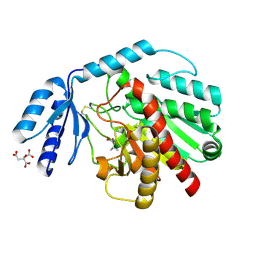 | | THREE DIMENSIONAL STRUCTURE OF PORCINE PANCREATIC PROCARBOXYPEPTIDASE A. A COMPARISON OF THE A AND B ZYMOGENS AND THEIR DETERMINANTS FOR INHIBITION AND ACTIVATION | | Descriptor: | CITRIC ACID, PROCARBOXYPEPTIDASE A PCPA, VALINE, ... | | Authors: | Guasch, A, Coll, M, Aviles, F.X, Huber, R. | | Deposit date: | 1991-10-28 | | Release date: | 1993-10-31 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Three-dimensional structure of porcine pancreatic procarboxypeptidase A. A comparison of the A and B zymogens and their determinants for inhibition and activation.
J.Mol.Biol., 224, 1992
|
|
1X7T
 
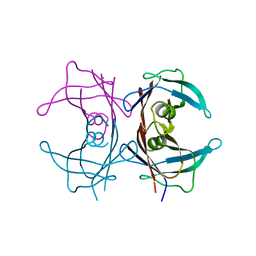 | | Structure of TTR R104H: a non-amyloidogenic variant with protective clinical effects | | Descriptor: | Transthyretin | | Authors: | Neto-Silva, R.M, Macedo-Ribeiro, S, Pereira, P.J.B, Coll, M, Saraiva, M.J, Damas, A.M. | | Deposit date: | 2004-08-16 | | Release date: | 2005-03-22 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | X-ray crystallographic studies of two transthyretin variants: further insights into amyloidogenesis.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
1SAX
 
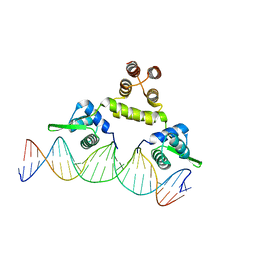 | | Three-dimensional structure of s.aureus methicillin-resistance regulating transcriptional repressor meci in complex with 25-bp ds-DNA | | Descriptor: | 5'-d(CAAAATTACAACTGTAATATCGGAG)-3', 5'-d(GCTCCGATATTACAGTTGTAATTTT)-3', Methicillin resistance regulatory protein mecI, ... | | Authors: | Garcia-Castellanos, R, Mallorqui-Fernandez, G, Marrero, A, Potempa, J, Coll, M, Gomis-Ruth, F.X. | | Deposit date: | 2004-02-09 | | Release date: | 2004-04-27 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | On the transcriptional regulation of methicillin resistance: MecI repressor in complex with its operator
J.Biol.Chem., 279, 2004
|
|
1RNF
 
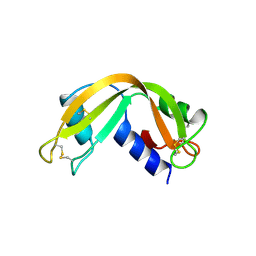 | | X-RAY CRYSTAL STRUCTURE OF UNLIGANDED HUMAN RIBONUCLEASE 4 | | Descriptor: | PROTEIN (RIBONUCLEASE 4) | | Authors: | Terzyan, S.S, Peracaula, R, De Llorens, R, Tsushima, Y, Yamada, H, Seno, M, Gomis-Rueth, F.X, Coll, M. | | Deposit date: | 1998-10-29 | | Release date: | 1999-10-29 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The three-dimensional structure of human RNase 4, unliganded and complexed with d(Up), reveals the basis for its uridine selectivity.
J.Mol.Biol., 285, 1999
|
|
1ZM5
 
 | | Conjugative Relaxase TRWC in complex with ORIT dna, cooper-bound structure | | Descriptor: | COPPER (II) ION, DNA (25-MER), SULFATE ION, ... | | Authors: | Boer, R, Russi, S, Guasch, A, Lucas, M, Blanco, A.G, Coll, M, de la Cruz, F. | | Deposit date: | 2005-05-10 | | Release date: | 2006-04-25 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Unveiling the Molecular Mechanism of a Conjugative Relaxase: The Structure of TrwC Complexed with a 27-mer DNA Comprising the Recognition Hairpin and the Cleavage Site
J.Mol.Biol., 358, 2006
|
|
1Z3F
 
 | | Structure of ellipticine in complex with a 6-bp DNA | | Descriptor: | 5'-D(*CP*GP*AP*TP*CP*G)-3', COBALT (II) ION, ELLIPTICINE | | Authors: | Canals, A, Purciolas, M, Aymami, J, Coll, M. | | Deposit date: | 2005-03-12 | | Release date: | 2005-07-19 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The anticancer agent ellipticine unwinds DNA by intercalative binding in an orientation parallel to base pairs.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
1NQ6
 
 | | Crystal Structure of the catalytic domain of xylanase A from Streptomyces halstedii JM8 | | Descriptor: | MAGNESIUM ION, Xys1 | | Authors: | Canals, A, Vega, M.C, Gomis-Ruth, F.X, Santamaria, R.I, Coll, M. | | Deposit date: | 2003-01-21 | | Release date: | 2004-01-21 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Structure of xylanase Xys1delta from Streptomyces halstedii.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
1DZA
 
 | | 3-D structure of a HP-RNase | | Descriptor: | RIBONUCLEASE 1 | | Authors: | Pous, J, Canals, A, Terzyan, S.S, Guasch, A, Benito, A, Ribo, M, Vilanova, M, Coll, M. | | Deposit date: | 2000-02-21 | | Release date: | 2001-02-16 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Three-Dimensional Structure of a Human Pancreatic Ribonuclease Variant, a Step Forward in the Design of Cytotoxic Ribonucleases
J.Mol.Biol., 303, 2000
|
|
4LDU
 
 | | Crystal structure of the DNA binding domain of Arabidopsis thaliana auxin response factor 5 | | Descriptor: | Auxin response factor 5, CHLORIDE ION | | Authors: | Boer, D.R, Freire-Rios, A, van den Berg, W.M.A, Weijers, D, Coll, M. | | Deposit date: | 2013-06-25 | | Release date: | 2014-02-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural Basis for DNA Binding Specificity by the Auxin-Dependent ARF Transcription Factors.
Cell(Cambridge,Mass.), 156, 2014
|
|
4LDW
 
 | |
4LDY
 
 | | Crystal structure of the DNA binding domain of the G245A mutant of arabidopsis thaliana auxin reponse factor 1 | | Descriptor: | Auxin response factor 1, CHLORIDE ION | | Authors: | Boer, D.R, Freire-Rios, A, van den Berg, W.M.A, Weijers, D, Coll, M. | | Deposit date: | 2013-06-25 | | Release date: | 2014-02-12 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Basis for DNA Binding Specificity by the Auxin-Dependent ARF Transcription Factors.
Cell(Cambridge,Mass.), 156, 2014
|
|
4LDV
 
 | | Crystal structure of the DNA binding domain of A. thailana auxin response factor 1 | | Descriptor: | Auxin response factor 1, CHLORIDE ION, FORMIC ACID, ... | | Authors: | boer, D.R, Freire-Rios, A, van den Berg, W.M.A, Weijers, D, Coll, M. | | Deposit date: | 2013-06-25 | | Release date: | 2014-02-12 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structural Basis for DNA Binding Specificity by the Auxin-Dependent ARF Transcription Factors.
Cell(Cambridge,Mass.), 156, 2014
|
|
4LDX
 
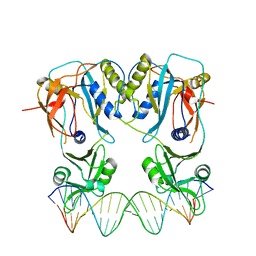 | | Crystal structure of the DNA binding domain of arabidopsis thaliana auxin response factor 1 (ARF1) in complex with protomor-like sequence ER7 | | Descriptor: | Auxin response factor 1, ER7, forward sequence, ... | | Authors: | Boer, D.R, Freire-Rios, A, van den Berg, W.M.A, Weijers, D, Coll, M. | | Deposit date: | 2013-06-25 | | Release date: | 2014-02-12 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural Basis for DNA Binding Specificity by the Auxin-Dependent ARF Transcription Factors.
Cell(Cambridge,Mass.), 156, 2014
|
|
1GL6
 
 | | Plasmid coupling protein TrwB in complex with the non-hydrolysable GTP analogue GDPNP | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ATPASE, CHLORIDE ION, ... | | Authors: | Gomis-Ruth, F.X, Moncalian, G, De La cruz, F, Coll, M. | | Deposit date: | 2001-08-28 | | Release date: | 2002-05-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The Bacterial Conjugation Protein Trwb Resembles Ring Helicases and F1-ATPase
Nature, 409, 2001
|
|
1GL7
 
 | | Plasmid coupling protein TrwB in complex with the non-hydrolisable ATP-analogue ADPNP. | | Descriptor: | CHLORIDE ION, CONJUGAL TRANSFER PROTEIN TRWB, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Gomis-Ruth, F.X, Moncalian, G, De La cruz, F, Coll, M. | | Deposit date: | 2001-08-28 | | Release date: | 2002-05-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The Bacterial Conjugation Protein Trwb Resembles Ring Helicases and F1-ATPase
Nature, 409, 2001
|
|
1GKI
 
 | | Plasmid coupling protein TrwB in complex with ADP and Mg2+. | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ADENOSINE-5'-DIPHOSPHATE, CONJUGAL TRANSFER PROTEIN TRWB, ... | | Authors: | Gomis-Ruth, F.X, Moncalian, G, De La cruz, F, Coll, M. | | Deposit date: | 2001-08-14 | | Release date: | 2002-05-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The Bacterial Conjugation Protein Trwb Resembles Ring Helicases and F1-ATPase
Nature, 409, 2001
|
|
1GVL
 
 | | Human prokallikrein 6 (hK6)/ prozyme/ proprotease M/ proneurosin | | Descriptor: | KALLIKREIN 6 | | Authors: | Gomis-Ruth, F.X, Bayes, A, Sotiropoulou, G, Pampalakis, G, Tsetsenis, T, Villegas, V, Aviles, F.X, Coll, M. | | Deposit date: | 2002-02-14 | | Release date: | 2002-05-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Structure of Human Prokallikrein 6 Reveals a Novel Activation Mechanism for the Kallikrein Family.
J.Biol.Chem., 277, 2002
|
|
1HE2
 
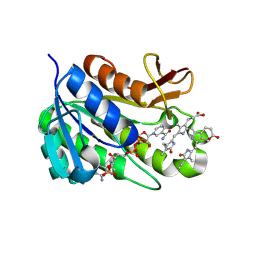 | | Human biliverdin IX beta reductase: NADP/biliverdin IX alpha ternary complex | | Descriptor: | BILIVERDIN IX BETA REDUCTASE, BILIVERDINE IX ALPHA, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Pereira, P.J.B, Macedo-Ribeiro, S, Parraga, A, Perez-Luque, R, Cunningham, O, Darcy, K, Mantle, T.J, Coll, M. | | Deposit date: | 2000-11-18 | | Release date: | 2001-02-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structure of Human Biliverdin Ix Beta Reductase, an Early Fetal Bilirubin Ix Producing Enzyme
Nat.Struct.Biol., 8, 2001
|
|
1HDO
 
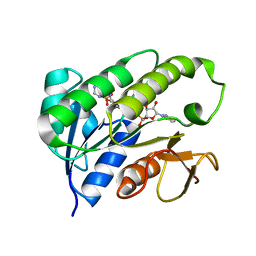 | | Human biliverdin IX beta reductase: NADP complex | | Descriptor: | BILIVERDIN IX BETA REDUCTASE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Pereira, P.J.B, Macedo-Ribeiro, S, Parraga, A, Perez-Luque, R, Cunningham, O, Darcy, K, Mantle, T.J, Coll, M. | | Deposit date: | 2000-11-16 | | Release date: | 2001-02-28 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Structure of Human Biliverdin Ix Beta Reductase, an Early Fetal Bilirubin Ix Producing Enzyme
Nat.Struct.Biol., 8, 2001
|
|
1E21
 
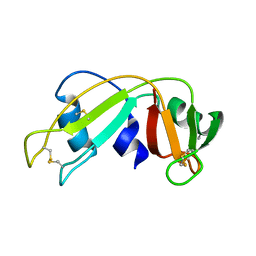 | | Ribonuclease 1 des1-7 Crystal Structure at 1.9A | | Descriptor: | RIBONUCLEASE 1 | | Authors: | Pous, J, Mallorqui-Fernandez, G, Peracaula, R, Terzyan, S.S, Futami, J, Tada, H, Yamada, H, Seno, M, De Llorens, R, Gomis-Ruth, F.X, Coll, M. | | Deposit date: | 2000-05-15 | | Release date: | 2001-05-03 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Three-Dimensional Crystal Structure of Human Rnase 1Dn7 at 1.9A Resolution
Acta Crystallogr.,Sect.D, 57, 2001
|
|
1DYT
 
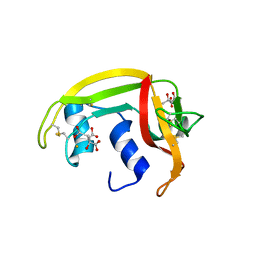 | | X-ray crystal structure of ECP (RNase 3) at 1.75 A | | Descriptor: | CITRIC ACID, EOSINOPHIL CATIONIC PROTEIN, FE (III) ION | | Authors: | Mallorqui-Fernandez, G, Pous, J, Peracaula, R, Maeda, T, Tada, H, Yamada, H, Seno, M, De Llorens, R, Gomis-Rueth, F.X, Coll, M. | | Deposit date: | 2000-02-08 | | Release date: | 2001-02-08 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Three-Dimensional Crystal Structure of Human Eosinophil Cationic Protein (Rnase 3) at 1.75 A Resolution.
J.Mol.Biol., 300, 2000
|
|
