8UYP
 
 | | SARS-CoV-1 5' proximal stem-loop 5 | | Descriptor: | SARS-CoV-1 5' proximal stem-loop 5 | | Authors: | Kretsch, R.C, Xu, L, Zheludev, I.N, Zhou, X, Huang, R, Nye, G, Li, S, Zhang, K, Chiu, W, Das, R. | | Deposit date: | 2023-11-13 | | Release date: | 2023-12-20 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (7.1 Å) | | Cite: | Tertiary folds of the SL5 RNA from the 5' proximal region of SARS-CoV-2 and related coronaviruses.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
5UU5
 
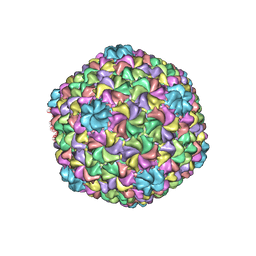 | | Bacteriophage P22 mature virion capsid protein | | Descriptor: | Major capsid protein | | Authors: | Hryc, C.F, Chen, D.-H, Afonine, P.V, Jakana, J, Wang, Z, Haase-Pettingell, C, Jiang, W, Adams, P.D, King, J.A, Schmid, M.F, Chiu, W. | | Deposit date: | 2017-02-16 | | Release date: | 2017-03-15 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Accurate model annotation of a near-atomic resolution cryo-EM map.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
6POM
 
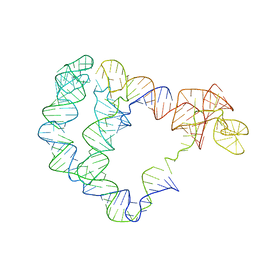 | | Cryo-EM structure of the full-length Bacillus subtilis glyQS T-box riboswitch in complex with tRNA-Gly | | Descriptor: | T-box GlyQS leader (155-MER), tRNAGly (75-MER) | | Authors: | Li, S, Su, Z, Zhang, J, Chiu, W. | | Deposit date: | 2019-07-04 | | Release date: | 2019-11-20 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4.9 Å) | | Cite: | Structural basis of amino acid surveillance by higher-order tRNA-mRNA interactions.
Nat.Struct.Mol.Biol., 26, 2019
|
|
1QVR
 
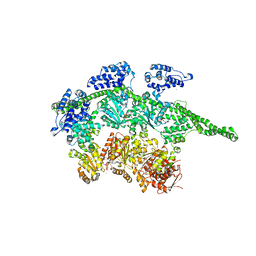 | | Crystal Structure Analysis of ClpB | | Descriptor: | ClpB protein, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, PLATINUM (II) ION | | Authors: | Lee, S, Sowa, M.E, Watanabe, Y, Sigler, P.B, Chiu, W, Yoshida, M, Tsai, F.T.F. | | Deposit date: | 2003-08-28 | | Release date: | 2003-10-21 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The Structure of ClpB: A Molecular Chaperone that Rescues Proteins from an Aggregated State
Cell(Cambridge,Mass.), 115, 2003
|
|
8TA5
 
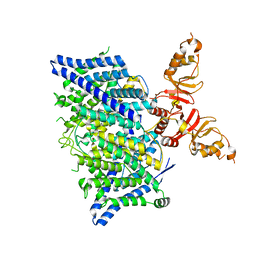 | | Title: Cryo-EM structure of the human CLC-2 chloride channel transmembrane domain with asymmetric C-terminal | | Descriptor: | Chloride channel protein 2 | | Authors: | Xu, M, Neelands, T, Powers, A.S, Liu, Y, Miller, S, Pintilie, G, Du Bois, J, Dror, R.O, Chiu, W, Maduke, M. | | Deposit date: | 2023-06-26 | | Release date: | 2024-01-31 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (2.76 Å) | | Cite: | CryoEM structures of the human CLC-2 voltage-gated chloride channel reveal a ball-and-chain gating mechanism.
Elife, 12, 2024
|
|
8TA4
 
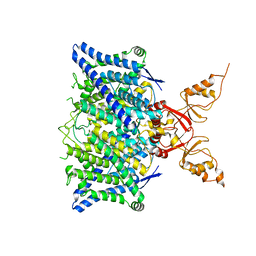 | | Cryo-EM structure of the human CLC-2 chloride channel transmembrane domain with symmetric C-terminal | | Descriptor: | CHLORIDE ION, Chloride channel protein 2 | | Authors: | Xu, M, Neelands, T, Powers, A.S, Liu, Y, Miller, S, Pintilie, G, Du Bois, J, Dror, R.O, Chiu, W, Maduke, M. | | Deposit date: | 2023-06-26 | | Release date: | 2024-01-31 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (2.75 Å) | | Cite: | CryoEM structures of the human CLC-2 voltage-gated chloride channel reveal a ball-and-chain gating mechanism.
Elife, 12, 2024
|
|
8TA2
 
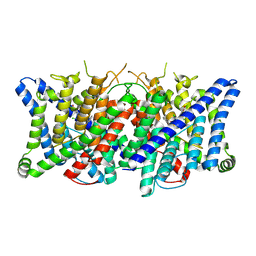 | | Cryo-EM structure of the human CLC-2 chloride channel transmembrane domain with bound inhibitor AK-42 | | Descriptor: | 2-[[2,6-bis(chloranyl)-3-phenylmethoxy-phenyl]amino]pyridine-3-carboxylic acid, CHLORIDE ION, Chloride channel protein 2 | | Authors: | Xu, M, Neelands, T, Powers, A.S, Liu, Y, Miller, S, Pintilie, G, Du Bois, J, Dror, R.O, Chiu, W, Maduke, M. | | Deposit date: | 2023-06-26 | | Release date: | 2024-01-31 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (2.74 Å) | | Cite: | CryoEM structures of the human CLC-2 voltage-gated chloride channel reveal a ball-and-chain gating mechanism.
Elife, 12, 2024
|
|
8TA6
 
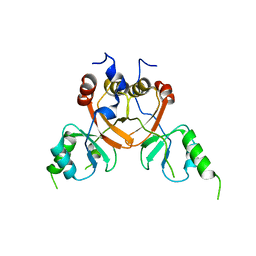 | | Cryo-EM structure of the human CLC-2 chloride channel C-terminal domain | | Descriptor: | Chloride channel protein 2 | | Authors: | Xu, M, Neelands, T, Powers, A.S, Liu, Y, Miller, S, Pintilie, G, Du Bois, J, Dror, R.O, Chiu, W, Maduke, M. | | Deposit date: | 2023-06-26 | | Release date: | 2024-01-31 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (4.03 Å) | | Cite: | CryoEM structures of the human CLC-2 voltage-gated chloride channel reveal a ball-and-chain gating mechanism.
Elife, 12, 2024
|
|
8TA3
 
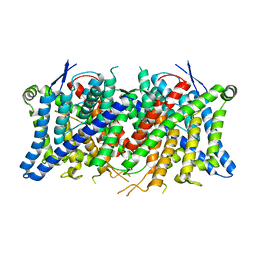 | | Cryo-EM structure of the human CLC-2 chloride channel transmembrane domain Apo state with resolved N-terminal hairpin | | Descriptor: | CHLORIDE ION, Chloride channel protein 2 | | Authors: | Xu, M, Neelands, T, Powers, A.S, Liu, Y, Miller, S, Pintilie, G, Du Bois, J, Dror, R.O, Chiu, W, Maduke, M. | | Deposit date: | 2023-06-26 | | Release date: | 2024-01-31 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (2.46 Å) | | Cite: | CryoEM structures of the human CLC-2 voltage-gated chloride channel reveal a ball-and-chain gating mechanism.
Elife, 12, 2024
|
|
1TT9
 
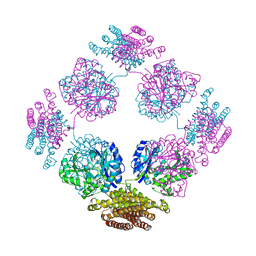 | | Structure of the bifunctional and Golgi associated formiminotransferase cyclodeaminase octamer | | Descriptor: | Formimidoyltransferase-cyclodeaminase (Formiminotransferase- cyclodeaminase) (FTCD) (58 kDa microtubule-binding protein) | | Authors: | Mao, Y, Vyas, N.K, Vyas, M.N, Chen, D.H, Ludtke, S.J, Chiu, W, Quiocho, F.A. | | Deposit date: | 2004-06-22 | | Release date: | 2005-06-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.42 Å) | | Cite: | Structure of the bifunctional and Golgi-associated formiminotransferase cyclodeaminase octamer
Embo J., 23, 2004
|
|
8FCG
 
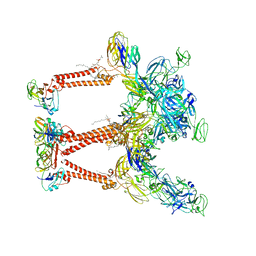 | | Cryo-EM structure of Chikungunya virus asymmetric unit | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Capsid protein, E1 glycoprotein, ... | | Authors: | Su, G.C, Chmielewsk, D, Kaelber, J, Pintilie, G, Chen, M, Jin, J, Auguste, A, Chiu, W. | | Deposit date: | 2022-12-01 | | Release date: | 2024-03-20 | | Last modified: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (3.09 Å) | | Cite: | Cryogenic electron microscopy and tomography reveal imperfect icosahedral symmetry in alphaviruses.
Pnas Nexus, 3, 2024
|
|
8FR7
 
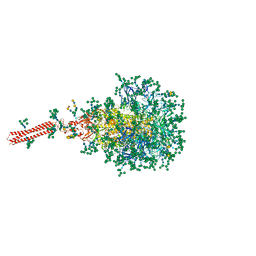 | | A hinge glycan regulates spike bending and impacts coronavirus infectivity | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-2)-[alpha-D-mannopyranose-(1-3)]alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Pintilie, G, Wilson, E, Chmielewski, D, Schmid, M.F, Jin, J, Chen, M, Singharoy, A, Chiu, W. | | Deposit date: | 2023-01-06 | | Release date: | 2023-10-04 | | Method: | ELECTRON MICROSCOPY (3.39 Å) | | Cite: | A hinge glycan regulates spike bending and impacts coronavirus infectivity
To Be Published
|
|
3ZQ1
 
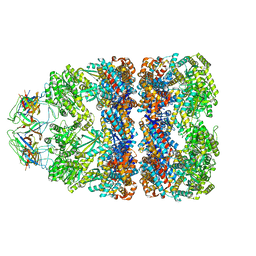 | | Visualizing GroEL-ES in the Act of Encapsulating a Non-Native Substrate Protein | | Descriptor: | 10 KDA CHAPERONIN, 60 KDA CHAPERONIN, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Chen, D.-H, Madan, D, Weaver, J, Lin, Z, Schroder, G.F, Chiu, W, Rye, H.S. | | Deposit date: | 2013-03-04 | | Release date: | 2013-06-19 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (15.9 Å) | | Cite: | Visualizing Groel/Es in the Act of Encapsulating a Folding Protein
Cell(Cambridge,Mass.), 153, 2013
|
|
3ZQ0
 
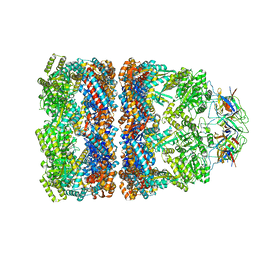 | | Visualizing GroEL-ES in the Act of Encapsulating a Non-Native Substrate Protein | | Descriptor: | 10 KDA CHAPERONIN, 60 KDA CHAPERONIN, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Chen, D.-H, Madan, D, Weaver, J, Lin, Z, Schroder, G.F, Chiu, W, Rye, H.S. | | Deposit date: | 2013-03-04 | | Release date: | 2013-06-19 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (9.2 Å) | | Cite: | Visualizing Groel/Es in the Act of Encapsulating a Folding Protein
Cell(Cambridge,Mass.), 153, 2013
|
|
3ZPZ
 
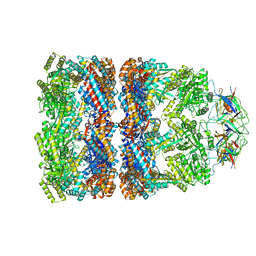 | | Visualizing GroEL-ES in the Act of Encapsulating a Non-Native Substrate Protein | | Descriptor: | 10 KDA CHAPERONIN, 60 KDA CHAPERONIN, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Chen, D.-H, Madan, D, Weaver, J, Lin, Z, Schroder, G.F, Chiu, W, Rye, H.S. | | Deposit date: | 2013-03-04 | | Release date: | 2013-06-19 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (8.9 Å) | | Cite: | Visualizing Groel/Es in the Act of Encapsulating a Folding Protein
Cell(Cambridge,Mass.), 153, 2013
|
|
4A0O
 
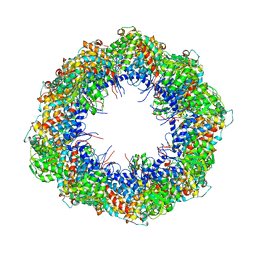 | | Symmetry-free cryo-EM map of TRiC in the nucleotide-free (apo) state | | Descriptor: | T-COMPLEX PROTEIN 1 SUBUNIT BETA | | Authors: | Cong, Y, Schroder, G.F, Meyer, A.S, Jakana, J, Ma, B, Dougherty, M.T, Schmid, M.F, Reissmann, S, Levitt, M, Ludtke, S.L, Frydman, J, Chiu, W. | | Deposit date: | 2011-09-10 | | Release date: | 2012-02-15 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (10.5 Å) | | Cite: | Symmetry-Free Cryo-Em Structures of the Chaperonin Tric Along its ATPase-Driven Conformational Cycle.
Embo J., 31, 2012
|
|
4A0W
 
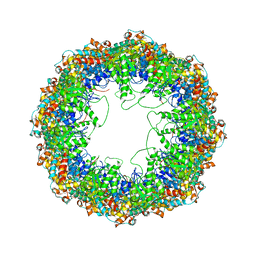 | | model built against symmetry-free cryo-EM map of TRiC-ADP-AlFx | | Descriptor: | T-COMPLEX PROTEIN 1 SUBUNIT BETA | | Authors: | Cong, Y, Schroder, G.F, Meyer, A.S, Jakana, J, Ma, B, Dougherty, M.T, Schmid, M.F, Reissmann, S, Levitt, M, Ludtke, S.L, Frydman, J, Chiu, W. | | Deposit date: | 2011-09-13 | | Release date: | 2012-02-15 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (13.9 Å) | | Cite: | Symmetry-Free Cryo-Em Structures of the Chaperonin Tric Along its ATPase-Driven Conformational Cycle.
Embo J., 31, 2012
|
|
4A13
 
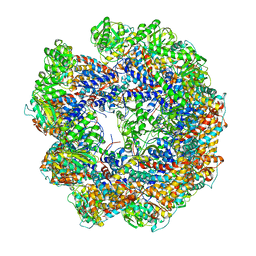 | | model refined against symmetry-free cryo-EM map of TRiC-ADP | | Descriptor: | T-COMPLEX PROTEIN 1 SUBUNIT BETA | | Authors: | Cong, Y, Schroder, G.F, Meyer, A.S, Jakana, J, Ma, B, Dougherty, M.T, Schmid, M.F, Reissmann, S, Levitt, M, Ludtke, S.L, Frydman, J, Chiu, W. | | Deposit date: | 2011-09-13 | | Release date: | 2012-02-15 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (11.3 Å) | | Cite: | Symmetry-Free Cryo-Em Structures of the Chaperonin Tric Along its ATPase-Driven Conformational Cycle.
Embo J., 31, 2012
|
|
4A0V
 
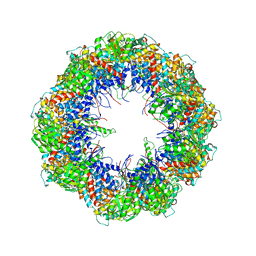 | | model refined against the Symmetry-free cryo-EM map of TRiC-AMP-PNP | | Descriptor: | T-COMPLEX PROTEIN 1 SUBUNIT BETA | | Authors: | Cong, Y, Schroder, G.F, Meyer, A.S, Jakana, J, Ma, B, Dougherty, M.T, Schmid, M.F, Reissmann, S, Levitt, M, Ludtke, S.L, Frydman, J, Chiu, W. | | Deposit date: | 2011-09-13 | | Release date: | 2012-02-15 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (10.7 Å) | | Cite: | Symmetry-Free Cryo-Em Structures of the Chaperonin Tric Along its ATPase-Driven Conformational Cycle.
Embo J., 31, 2012
|
|
7RRP
 
 | |
4BML
 
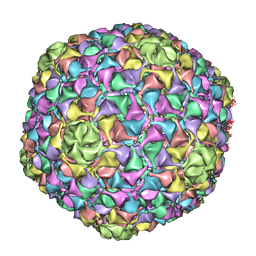 | | C-alpha backbone trace of major capsid protein gp39 found in marine virus Syn5. | | Descriptor: | MAJOR CAPSID PROTEIN | | Authors: | Gipson, P, Baker, M.L, Raytcheva, D, Haase-Pettingell, C, Piret, J, King, J, Chiu, W. | | Deposit date: | 2013-05-09 | | Release date: | 2014-05-21 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | Protruding Knob-Like Proteins Violate Local Symmetries in an Icosahedral Marine Virus.
Nat.Commun., 5, 2014
|
|
2XD8
 
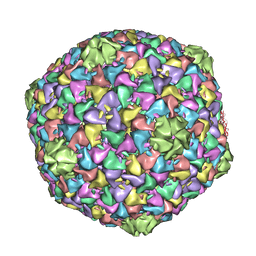 | | Capsid structure of the infectious Prochlorococcus Cyanophage P-SSP7 | | Descriptor: | T7-LIKE CAPSID PROTEIN | | Authors: | Liu, X, Zhang, Q, Murata, K, Baker, M.L, Sullivan, M.B, Fu, C, Dougherty, M, Schmid, M.F, Osburne, M.S, Chisholm, S.W, Chiu, W. | | Deposit date: | 2010-04-30 | | Release date: | 2010-06-16 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Structural Changes in a Marine Podovirus Associated with Release of its Genome Into Prochlorococcus
Nat.Struct.Mol.Biol., 17, 2010
|
|
2YEW
 
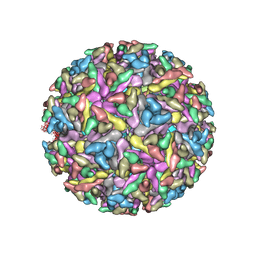 | | Modeling Barmah Forest virus structural proteins | | Descriptor: | CAPSID PROTEIN, E1 ENVELOPE GLYCOPROTEIN, E2 ENVELOPE GLYCOPROTEIN | | Authors: | Kostyuchenko, V.A, Jakana, J, Liu, X, Haddow, A.D, Aung, M, Weaver, S.C, Chiu, W, Lok, S.M. | | Deposit date: | 2011-03-31 | | Release date: | 2012-04-18 | | Last modified: | 2019-10-23 | | Method: | ELECTRON MICROSCOPY (5 Å) | | Cite: | The Structure of Barmah Forest Virus as Revealed by Cryo-Electron Microscopy at a 6-Angstrom Resolution Has Detailed Transmembrane Protein Architecture and Interactions.
J.Virol., 85, 2011
|
|
2L1F
 
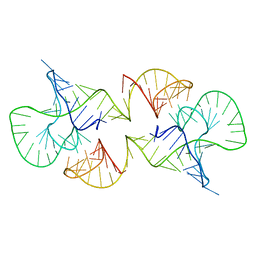 | | Structure of a conserved retroviral RNA packaging element by NMR spectroscopy and cryo-electron tomography | | Descriptor: | RNA (65-MER), RNA (66-MER) | | Authors: | Summers, M.F, Irobalieva, R.N, Tolbert, B, Smalls-Manty, A, Iyalla, K, Loeliger, K, D'Souza, V, Khant, H, Schmid, M, Garcia, E, Telesnitsky, A, Chiu, W, Miyazaki, Y. | | Deposit date: | 2010-07-28 | | Release date: | 2010-10-27 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure of a conserved retroviral RNA packaging element by NMR spectroscopy and cryo-electron tomography.
J.Mol.Biol., 404, 2010
|
|
2M5S
 
 | | High-resolution NMR structure and cryo-EM imaging support multiple functional roles for the accessory I-domain of phage P22 coat protein | | Descriptor: | Coat protein | | Authors: | Rizzo, A.A, Suhanovsky, M.M, Baker, M.L, Fraser, L.C.R, Jones, L.M, Rempel, D.L, Gross, M.L, Chiu, W, Alexandrescu, A.T, Teschke, C.M. | | Deposit date: | 2013-03-05 | | Release date: | 2014-03-05 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Multiple Functional Roles of the Accessory I-Domain of Bacteriophage P22 Coat Protein Revealed by NMR Structure and CryoEM Modeling.
Structure, 22, 2014
|
|
