1PRM
 
 | | TWO BINDING ORIENTATIONS FOR PEPTIDES TO SRC SH3 DOMAIN: DEVELOPMENT OF A GENERAL MODEL FOR SH3-LIGAND INTERACTIONS | | Descriptor: | C-SRC TYROSINE KINASE SH3 DOMAIN, PROLINE-RICH LIGAND PLR1 (AFAPPLPRR) | | Authors: | Feng, S, Chen, J.K, Yu, H, Simon, J.A, Schreiber, S.L. | | Deposit date: | 1994-10-10 | | Release date: | 1995-02-07 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Two binding orientations for peptides to the Src SH3 domain: development of a general model for SH3-ligand interactions.
Science, 266, 1994
|
|
1PRL
 
 | | TWO BINDING ORIENTATIONS FOR PEPTIDES TO SRC SH3 DOMAIN: DEVELOPMENT OF A GENERAL MODEL FOR SH3-LIGAND INTERACTIONS | | Descriptor: | C-SRC TYROSINE KINASE SH3 DOMAIN, PROLINE-RICH LIGAND PLR1 (AFAPPLPRR) | | Authors: | Feng, S, Chen, J.K, Yu, H, Simon, J.A, Schreiber, S.L. | | Deposit date: | 1994-10-10 | | Release date: | 1995-02-07 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Two binding orientations for peptides to the Src SH3 domain: development of a general model for SH3-ligand interactions.
Science, 266, 1994
|
|
1PHT
 
 | | PHOSPHATIDYLINOSITOL 3-KINASE P85-ALPHA SUBUNIT SH3 DOMAIN, RESIDUES 1-85 | | Descriptor: | PHOSPHATIDYLINOSITOL 3-KINASE P85-ALPHA SUBUNIT | | Authors: | Liang, J, Chen, J.K, Schreiber, S.L, Clardy, J. | | Deposit date: | 1995-08-17 | | Release date: | 1995-12-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of P13K SH3 domain at 20 angstroms resolution.
J.Mol.Biol., 257, 1996
|
|
1RLQ
 
 | | TWO BINDING ORIENTATIONS FOR PEPTIDES TO SRC SH3 DOMAIN: DEVELOPMENT OF A GENERAL MODEL FOR SH3-LIGAND INTERACTIONS | | Descriptor: | C-SRC TYROSINE KINASE SH3 DOMAIN, PROLINE-RICH LIGAND RLP2 (RALPPLPRY) | | Authors: | Feng, S, Chen, J.K, Yu, H, Simon, J.A, Schreiber, S.L. | | Deposit date: | 1994-10-10 | | Release date: | 1995-02-07 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Two binding orientations for peptides to the Src SH3 domain: development of a general model for SH3-ligand interactions.
Science, 266, 1994
|
|
1RLP
 
 | | TWO BINDING ORIENTATIONS FOR PEPTIDES TO SRC SH3 DOMAIN: DEVELOPMENT OF A GENERAL MODEL FOR SH3-LIGAND INTERACTIONS | | Descriptor: | C-SRC TYROSINE KINASE SH3 DOMAIN, PROLINE-RICH LIGAND RLP2 (RALPPLPRY) | | Authors: | Feng, S, Chen, J.K, Yu, H, Simon, J.A, Schreiber, S.L. | | Deposit date: | 1994-10-10 | | Release date: | 1995-02-07 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Two binding orientations for peptides to the Src SH3 domain: development of a general model for SH3-ligand interactions.
Science, 266, 1994
|
|
6E08
 
 | |
6E07
 
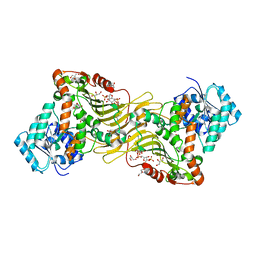 | | Crystal structure of Canton G6PD in complex with structural NADP | | Descriptor: | GLYCEROL, Glucose-6-phosphate 1-dehydrogenase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Rahighi, S, Mochly-Rosen, D, Wakatsuki, S. | | Deposit date: | 2018-07-06 | | Release date: | 2018-07-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Correcting glucose-6-phosphate dehydrogenase deficiency with a small-molecule activator.
Nat Commun, 9, 2018
|
|
1NLP
 
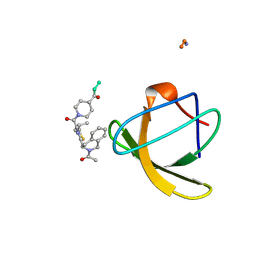 | | STRUCTURE OF SIGNAL TRANSDUCTION PROTEIN, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | C-SRC, NL2 (MN8-MN1-PLPPLP) | | Authors: | Feng, S, Kapoor, T.M, Shirai, F, Combs, A.P, Schreiber, S.L. | | Deposit date: | 1996-08-04 | | Release date: | 1997-01-27 | | Last modified: | 2023-11-15 | | Method: | SOLUTION NMR | | Cite: | Molecular basis for the binding of SH3 ligands with non-peptide elements identified by combinatorial synthesis.
Chem.Biol., 3, 1996
|
|
1NLO
 
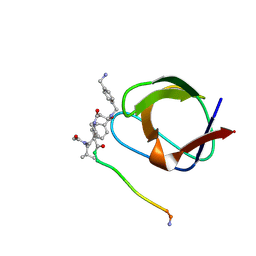 | | STRUCTURE OF SIGNAL TRANSDUCTION PROTEIN, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | C-SRC, NL1 (MN7-MN2-MN1-PLPPLP) | | Authors: | Feng, S, Kapoor, T.M, Shirai, F, Combs, A.P, Schreiber, S.L. | | Deposit date: | 1996-08-04 | | Release date: | 1997-01-27 | | Last modified: | 2023-11-15 | | Method: | SOLUTION NMR | | Cite: | Molecular basis for the binding of SH3 ligands with non-peptide elements identified by combinatorial synthesis.
Chem.Biol., 3, 1996
|
|
1QWE
 
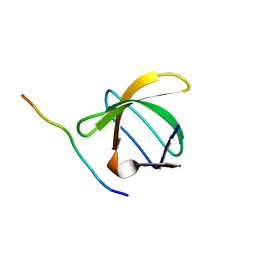 | | C-SRC SH3 DOMAIN COMPLEXED WITH LIGAND APP12 | | Descriptor: | ALA-PRO-PRO-LEU-PRO-PRO-ARG-ASN-ARG-PRO-ARG-LEU, TYROSINE-PROTEIN KINASE TRANSFORMING PROTEIN SRC | | Authors: | Feng, S, Chiyoshi, K, Rickles, R.J, Schreiber, S.L. | | Deposit date: | 1995-11-09 | | Release date: | 1996-03-08 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Specific interactions outside the proline-rich core of two classes of Src homology 3 ligands.
Proc.Natl.Acad.Sci.USA, 92, 1995
|
|
1QWF
 
 | | C-SRC SH3 DOMAIN COMPLEXED WITH LIGAND VSL12 | | Descriptor: | TYROSINE-PROTEIN KINASE TRANSFORMING PROTEIN SRC, VAL-SER-LEU-ALA-ARG-ARG-PRO-LEU-PRO-PRO-LEU-PRO | | Authors: | Feng, S, Chiyoshi, K, Rickles, R.J, Schreiber, S.L. | | Deposit date: | 1995-11-09 | | Release date: | 1996-03-08 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Specific interactions outside the proline-rich core of two classes of Src homology 3 ligands.
Proc.Natl.Acad.Sci.USA, 92, 1995
|
|
4DOV
 
 | | Structure of free mouse ORC1 BAH domain | | Descriptor: | Origin recognition complex subunit 1 | | Authors: | Song, J, Patel, D.J. | | Deposit date: | 2012-02-10 | | Release date: | 2012-03-07 | | Last modified: | 2012-04-11 | | Method: | X-RAY DIFFRACTION (1.696 Å) | | Cite: | The BAH domain of ORC1 links H4K20me2 to DNA replication licensing and Meier-Gorlin syndrome.
Nature, 484, 2012
|
|
4DOW
 
 | | Structure of mouse ORC1 BAH domain bound to H4K20me2 | | Descriptor: | Histone H4, Origin recognition complex subunit 1 | | Authors: | Song, J, Patel, D.J. | | Deposit date: | 2012-02-10 | | Release date: | 2012-03-07 | | Last modified: | 2012-04-11 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The BAH domain of ORC1 links H4K20me2 to DNA replication licensing and Meier-Gorlin syndrome.
Nature, 484, 2012
|
|
7MJD
 
 | | Crystal Structure Analysis of ALDH1B1 | | Descriptor: | 8-(2-methoxyphenyl)-10-(4-phenylphenyl)-1$l^{4},8-diazabicyclo[5.3.0]deca-1(7),9-diene, Aldehyde dehydrogenase X, mitochondrial, ... | | Authors: | Fernandez, D, Chen, J.K. | | Deposit date: | 2021-04-20 | | Release date: | 2022-06-08 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Inhibitors targeted to aldehyde dehydrogenase
Nat.Chem.Biol., 2022
|
|
7MJC
 
 | | Crystal Structure Analysis of ALDH1B1 | | Descriptor: | Aldehyde dehydrogenase X, mitochondrial, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Fernandez, D, Chen, J.K. | | Deposit date: | 2021-04-20 | | Release date: | 2022-06-08 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Inhibitors targeted to aldehyde dehydrogenase
Nat.Chem.Biol., 2022
|
|
7RAD
 
 | | Crystal Structure Analysis of ALDH1B1 | | Descriptor: | 1,2-ETHANEDIOL, 3-(2-methoxyphenyl)-1-(4-phenylphenyl)-6,7,8,9-tetrahydro-5~{H}-imidazo[1,2-a][1,3]diazepine, Aldehyde dehydrogenase X, ... | | Authors: | Fernandez, D, Chen, J.K. | | Deposit date: | 2021-07-01 | | Release date: | 2022-06-08 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Inhibitors targeted to aldehyde dehydrogenase
Nat.Chem.Biol., 2022
|
|
