5XS1
 
 | |
4IXA
 
 | | Structure of DNA-binding domain of the response regulator SaeR from Staphylococcus epidermidis | | Descriptor: | Response regulator SaeR | | Authors: | Chen, Y.R, Chen, S.C, Yang, C.S, Kuan, S.M, Liu, Y.H, Chen, Y. | | Deposit date: | 2013-01-24 | | Release date: | 2014-01-29 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure of DNA-binding domain of the response regulator SaeR from Staphylococcus epidermidis
To be Published
|
|
4JIS
 
 | | Crystal structure of ribitol 5-phosphate cytidylyltransferase (TarI) from Bacillus subtilis | | Descriptor: | ribitol-5-phosphate cytidylyltransferase | | Authors: | Yang, C.S, Chen, S.C, Chen, Y.R, Kuan, S.M, Liu, Y.H, Chen, Y. | | Deposit date: | 2013-03-06 | | Release date: | 2014-03-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.772 Å) | | Cite: | Crystal structure of ribitol 5-phosphate cytidylyltransferase (TarI) from Bacillus subtilis
To be Published
|
|
4FKZ
 
 | | Crystal structure of Bacillus subtilis UDP-GlcNAc 2-epimerase in complex with UDP-GlcNAc and UDP | | Descriptor: | UDP-N-acetylglucosamine 2-epimerase, URIDINE-5'-DIPHOSPHATE, URIDINE-DIPHOSPHATE-N-ACETYLGLUCOSAMINE | | Authors: | Yang, C.S, Chen, S.C, Kuan, S.M, Chen, Y.R, Liu, Y.H, Chen, Y. | | Deposit date: | 2012-06-14 | | Release date: | 2013-05-08 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Crystal structure of Bacillus subtilis UDP-GlcNAc 2-epimerase in complex with UDP-GlcNAc and UDP
To be Published
|
|
8W68
 
 | | Crystal structure of Q9PR55 at pH 6.0 (use NMR model) | | Descriptor: | Uncharacterized protein UU089.1 | | Authors: | Hsu, M.F, Ko, T.P, Huang, K.F, Chen, Y.R, Huang, J.S, Hsu, S.T.D. | | Deposit date: | 2023-08-28 | | Release date: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure, dynamics, and stability of the smallest and most complex 7 1 protein knot.
J.Biol.Chem., 300, 2023
|
|
8IWC
 
 | | Crystal structure of Q9PR55 at pH 6.0 | | Descriptor: | Uncharacterized protein UU089.1 | | Authors: | Hsu, M.F, Ko, T.P, Huang, K.F, Chen, Y.R, Huang, J.S, Hsu, S.T.D. | | Deposit date: | 2023-03-29 | | Release date: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Structure, dynamics, and stability of the smallest and most complex 7 1 protein knot.
J.Biol.Chem., 300, 2023
|
|
8IWA
 
 | | Crystal structure of Q9PR55 at pH 6.5 | | Descriptor: | SULFATE ION, Uncharacterized protein UU089.1 | | Authors: | Hsu, M.F, Ko, T.P, Huang, K.F, Chen, Y.R, Huang, J.S, Hsu, S.T.D. | | Deposit date: | 2023-03-29 | | Release date: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Structure, dynamics, and stability of the smallest and most complex 7 1 protein knot.
J.Biol.Chem., 300, 2023
|
|
8IWB
 
 | | Crystal structure of Q9PR55 at pH 7.5 | | Descriptor: | Uncharacterized protein UU089.1 | | Authors: | Hsu, M.F, Ko, T.P, Huang, K.F, Chen, Y.R, Huang, J.S, Hsu, S.T.D. | | Deposit date: | 2023-03-29 | | Release date: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Structure, dynamics, and stability of the smallest and most complex 7 1 protein knot.
J.Biol.Chem., 300, 2023
|
|
5MZP
 
 | | Crystal structure of stabilized A2A adenosine receptor A2AR-StaR2-bRIL in complex with caffeine at 2.1A resolution | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Adenosine receptor A2a,Soluble cytochrome b562,Adenosine receptor A2a, CAFFEINE, ... | | Authors: | Cheng, K.Y.R, Segala, E, Robertson, N, Deflorian, F, Dore, A.S, Errey, J.C, Fiez-Vandal, C, Marshall, F.H, Cooke, R.M. | | Deposit date: | 2017-02-01 | | Release date: | 2017-07-26 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structures of Human A1 and A2A Adenosine Receptors with Xanthines Reveal Determinants of Selectivity.
Structure, 25, 2017
|
|
8Z95
 
 | | Humanized anti-PEG h6.3 Fab in complex with PEG | | Descriptor: | 2,5,8,11,14,17,20,23,26,29,32,35,38,41,44,47,50,53,56,59,62,65,68,71,74,77,80-HEPTACOSAOXADOOCTACONTAN-82-OL, Heavy chain of Anti-PEG antibody h6-3 Fab fragment, ISOPROPYL ALCOHOL, ... | | Authors: | Lin, Y.C, Chang, C.Y, Su, Y.C. | | Deposit date: | 2024-04-22 | | Release date: | 2025-02-12 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | pH-Responsive Polyethylene Glycol Engagers for Enhanced Brain Delivery of PEGylated Nanomedicine to Treat Glioblastoma.
Acs Nano, 19, 2025
|
|
4IWM
 
 | | Crystal Structure of the Conserved Hypothetical Protein MJ0927 from Methanocaldococcus jannaschii (in P21 form) | | Descriptor: | UPF0135 protein MJ0927 | | Authors: | Kuan, S.M, Chen, S.C, Yang, C.S, Chen, Y.R, Liu, Y.H, Chen, Y. | | Deposit date: | 2013-01-24 | | Release date: | 2014-01-29 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of a conserved hypothetical protein MJ0927 from Methanocaldococcus jannaschii reveals a novel quaternary assembly in the Nif3 family.
Biomed Res Int, 2014, 2014
|
|
4IWG
 
 | | Crystal Structure of the Conserved Hypothetical Protein MJ0927 from Methanocaldococcus jannaschii (in C2221 form) | | Descriptor: | UPF0135 protein MJ0927 | | Authors: | Kuan, S.M, Chen, S.C, Yang, C.S, Chen, Y.R, Liu, Y.H, Chen, Y. | | Deposit date: | 2013-01-23 | | Release date: | 2014-01-29 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.472 Å) | | Cite: | Crystal structure of a conserved hypothetical protein MJ0927 from Methanocaldococcus jannaschii reveals a novel quaternary assembly in the Nif3 family.
Biomed Res Int, 2014, 2014
|
|
2N93
 
 | | Solution structure of lcFABP | | Descriptor: | Fatty acid-binding protein | | Authors: | Tseng, K, Lyu, P. | | Deposit date: | 2015-11-05 | | Release date: | 2016-01-20 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | 1H, 15N and 13C resonance assignments of light organ-associated fatty acid-binding protein of Taiwanese fireflies.
Biomol NMR Assign, 10, 2016
|
|
8K3Y
 
 | | The "5+1" heteromeric structure of Lon protease consisting of a spiral pentamer with Y224S mutation and an N-terminal-truncated monomeric E613K mutant | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Lon protease | | Authors: | Li, S, Hsieh, K.Y, Kuo, C.I, Zhang, K, Chang, C.I. | | Deposit date: | 2023-07-17 | | Release date: | 2023-10-25 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (4.42 Å) | | Cite: | A 5+1 assemble-to-activate mechanism of the Lon proteolytic machine.
Nat Commun, 14, 2023
|
|
7YPH
 
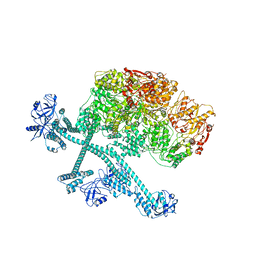 | | Open-spiral pentamer of the substrate-free Lon protease with a Y224S mutation | | Descriptor: | Lon protease, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER | | Authors: | Li, S, Hsieh, K.Y, Kuo, C.I, Lee, S.H, Ho, M.R, Wang, C.H, Zhang, K, Chang, C.I. | | Deposit date: | 2022-08-03 | | Release date: | 2023-10-25 | | Last modified: | 2023-11-29 | | Method: | ELECTRON MICROSCOPY (3.68 Å) | | Cite: | A 5+1 assemble-to-activate mechanism of the Lon proteolytic machine.
Nat Commun, 14, 2023
|
|
7YUT
 
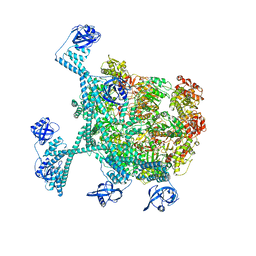 | | MtaLon-Apo for the spiral oligomers of hexamer | | Descriptor: | Lon protease | | Authors: | Li, S, Hsieh, K, Kuo, C, Lee, S, Ho, M, Wang, C, Zhang, K, Chang, C.I. | | Deposit date: | 2022-08-17 | | Release date: | 2023-10-25 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | A 5+1 assemble-to-activate mechanism of the Lon proteolytic machine.
Nat Commun, 14, 2023
|
|
7YUX
 
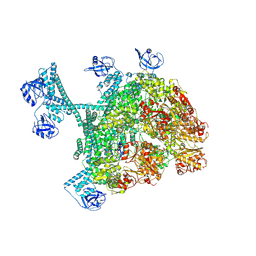 | | MtaLon-ADP for the spiral oligomers of hexamer | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Lon protease | | Authors: | Li, S, Hsieh, K, Kuo, C, Lee, S, Ho, M, Wang, C, Zhang, K, Chang, C.I. | | Deposit date: | 2022-08-18 | | Release date: | 2023-10-25 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | A 5+1 assemble-to-activate mechanism of the Lon proteolytic machine.
Nat Commun, 14, 2023
|
|
7YPJ
 
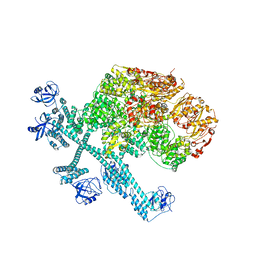 | | Spiral pentamer of the substrate-free Lon protease with a S678A mutation | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Lon protease | | Authors: | Li, S, Hsieh, K.Y, Kuo, C.I, Lee, S.H, Ho, M.R, Wang, C.H, Zhang, K, Chang, C.I. | | Deposit date: | 2022-08-03 | | Release date: | 2023-10-25 | | Last modified: | 2023-11-29 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | A 5+1 assemble-to-activate mechanism of the Lon proteolytic machine.
Nat Commun, 14, 2023
|
|
7YUU
 
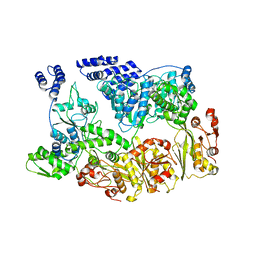 | | MtaLon-ADP for the spiral oligomers of trimer | | Descriptor: | Lon protease | | Authors: | Li, S, Hsieh, K, Kuo, C, Lee, S, Ho, M, Wang, C, Zhang, K, Chang, C.I. | | Deposit date: | 2022-08-17 | | Release date: | 2023-10-25 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (5.8 Å) | | Cite: | A 5+1 assemble-to-activate mechanism of the Lon proteolytic machine.
Nat Commun, 14, 2023
|
|
7YUV
 
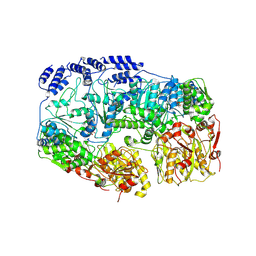 | | MtaLon-ADP for the spiral oligomers of tetramer | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Lon protease | | Authors: | Li, S, Hsieh, K, Kuo, C, Lee, S, Ho, M, Wang, C, Zhang, K, Chang, C.I. | | Deposit date: | 2022-08-17 | | Release date: | 2023-10-25 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | A 5+1 assemble-to-activate mechanism of the Lon proteolytic machine.
Nat Commun, 14, 2023
|
|
7YUW
 
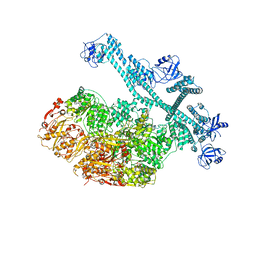 | | MtaLon-ADP for the spiral oligomers of pentamer | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Lon protease | | Authors: | Li, S, Hsieh, K, Kuo, C, Lee, S, Ho, M, Wang, C, Zhang, K, Chang, C.I. | | Deposit date: | 2022-08-18 | | Release date: | 2023-10-25 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | A 5+1 assemble-to-activate mechanism of the Lon proteolytic machine.
Nat Commun, 14, 2023
|
|
7YPK
 
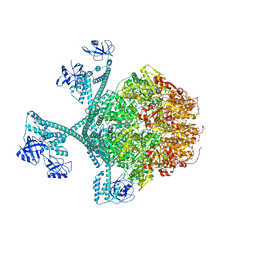 | | Close-ring hexamer of the substrate-bound Lon protease with an S678A mutation | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Lon protease, alpha-S1-casein | | Authors: | Li, S, Hsieh, K.Y, Kuo, C.I, Lee, S.H, Ho, M.R, Wang, C.H, Zhang, K, Chang, C.I. | | Deposit date: | 2022-08-03 | | Release date: | 2023-10-25 | | Last modified: | 2023-11-29 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | A 5+1 assemble-to-activate mechanism of the Lon proteolytic machine.
Nat Commun, 14, 2023
|
|
7YUM
 
 | | MtaLon-Apo for the spiral oligomers of tetramer | | Descriptor: | Lon protease | | Authors: | Li, S, Hsieh, K, Kuo, C, Lee, S, Ho, M, Wang, C, Zhang, K, Chang, C.I. | | Deposit date: | 2022-08-17 | | Release date: | 2023-10-25 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | A 5+1 assemble-to-activate mechanism of the Lon proteolytic machine.
Nat Commun, 14, 2023
|
|
7YPI
 
 | | Spiral hexamer of the substrate-free Lon protease with a Y224S mutation | | Descriptor: | Lon protease, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER | | Authors: | Li, S, Hsieh, K.Y, Kuo, C.I, Lee, S.H, Ho, M.R, Wang, C.H, Zhang, K, Chang, C.I. | | Deposit date: | 2022-08-03 | | Release date: | 2023-10-25 | | Last modified: | 2023-11-29 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | A 5+1 assemble-to-activate mechanism of the Lon proteolytic machine.
Nat Commun, 14, 2023
|
|
7YUH
 
 | | MtaLon-Apo for the spiral oligomers of trimer | | Descriptor: | Lon protease | | Authors: | Li, S, Hsieh, K, Kuo, C, Lee, S, Ho, M, Wang, C, Zhang, K, Chang, C.I. | | Deposit date: | 2022-08-17 | | Release date: | 2023-10-25 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | A 5+1 assemble-to-activate mechanism of the Lon proteolytic machine.
Nat Commun, 14, 2023
|
|
