4GN1
 
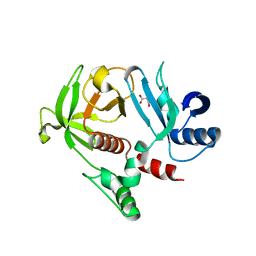 | | Crystal Structure of the RA and PH domains of Lamellipodin | | Descriptor: | MALONATE ION, Ras-associated and pleckstrin homology domains-containing protein 1 | | Authors: | Chang, Y.C.E, Wu, J. | | Deposit date: | 2012-08-16 | | Release date: | 2012-11-28 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of Lamellipodin implicates diverse functions in actin polymerization and Ras signaling.
Protein Cell, 4, 2013
|
|
8XMI
 
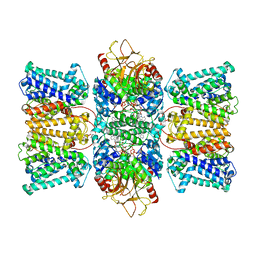 | | Potassium transporter KtrAB from Bacillus subtilis in ATP-bound state with addition of EDTA and EGTA, C1 symmetry | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Ktr system potassium uptake protein A, Ktr system potassium uptake protein B, ... | | Authors: | Chang, Y.K, Chiang, W.T, Hu, N.J, Tsai, M.D. | | Deposit date: | 2023-12-27 | | Release date: | 2024-04-03 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis and synergism of ATP and Na+ activation in bacterial K+ uptake system KtrAB
Nat Commun, 15, 2024
|
|
8XMH
 
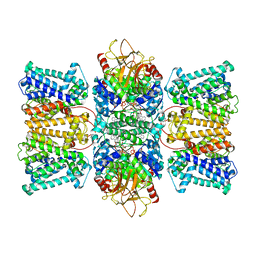 | | Potassium transporter KtrAB from Bacillus subtilis in ATP-bound state with addition of EDTA and EGTA, vertical C2 symmetry axis | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Ktr system potassium uptake protein A, Ktr system potassium uptake protein B, ... | | Authors: | Chang, Y.K, Chiang, W.T, Hu, N.J, Tsai, M.D. | | Deposit date: | 2023-12-27 | | Release date: | 2024-04-03 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (2.85 Å) | | Cite: | Structural basis and synergism of ATP and Na+ activation in bacterial K+ uptake system KtrAB
Nat Commun, 15, 2024
|
|
7QU8
 
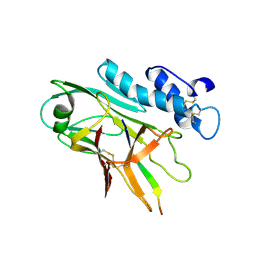 | | ADGRG3/GPR97 Extracellular Region | | Descriptor: | Adhesion G protein-coupled receptor G3 | | Authors: | Zheng-Gerard, C, Chu, T.Y, El Omari, K, Lin, H.H, Seiradake, E. | | Deposit date: | 2022-01-17 | | Release date: | 2022-09-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.37 Å) | | Cite: | GPR97-mediated PAR2 transactivation via a mPR3-associated macromolecular complex induces inflammatory activation of human neutrophils
Nat Commun, 2022
|
|
6JX7
 
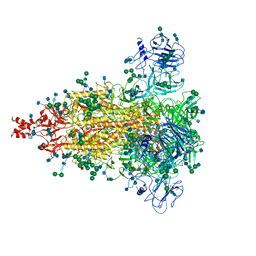 | | Cryo-EM structure of spike protein of feline infectious peritonitis virus strain UU4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Feline Infectious Peritonitis Virus Spike Protein, ... | | Authors: | Hsu, S.T.D, Yang, T.J, Ko, T.P, Draczkowski, P. | | Deposit date: | 2019-04-22 | | Release date: | 2020-01-15 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.31 Å) | | Cite: | Cryo-EM analysis of a feline coronavirus spike protein reveals a unique structure and camouflaging glycans.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
7W6M
 
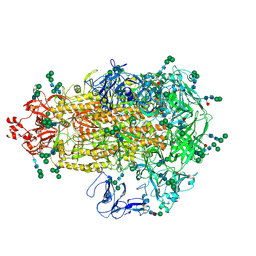 | | Cryo-EM map of PEDV (Pintung 52) S protein with all three protomers in the D0-down conformation determined in situ on intact viral particles. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Hsu, S.T.D, Draczkowski, P, Wang, Y.S. | | Deposit date: | 2021-12-02 | | Release date: | 2022-08-03 | | Last modified: | 2022-09-14 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | In situ structure and dynamics of an alphacoronavirus spike protein by cryo-ET and cryo-EM.
Nat Commun, 13, 2022
|
|
7W73
 
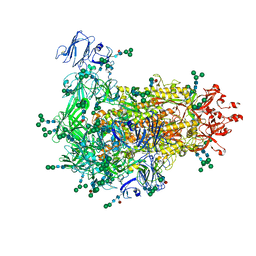 | | Cryo-EM map of PEDV S protein with one protomer in the D0-up conformation while the other two in the D0-down conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Hsu, S.T.D, Draczkowski, P, Wang, Y.S. | | Deposit date: | 2021-12-03 | | Release date: | 2022-08-03 | | Last modified: | 2022-12-21 | | Method: | ELECTRON MICROSCOPY (6.4 Å) | | Cite: | In situ structure and dynamics of an alphacoronavirus spike protein by cryo-ET and cryo-EM.
Nat Commun, 13, 2022
|
|
7Y6U
 
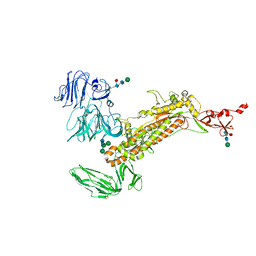 | | Symmetry-expanded and locally refined protomer structure of IPEC-J2 cell-derived PEDV PT52 S with a CTD-close conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Hsu, S.T.D, Draczkowski, P, Wang, Y.S. | | Deposit date: | 2022-06-21 | | Release date: | 2022-08-03 | | Last modified: | 2022-09-14 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | In situ structure and dynamics of an alphacoronavirus spike protein by cryo-ET and cryo-EM.
Nat Commun, 13, 2022
|
|
7Y6V
 
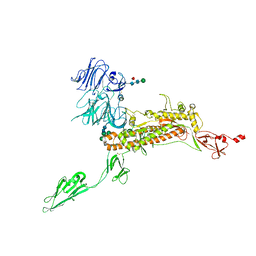 | | Symmetry-expanded and locally refined protomer structure of IPEC-J2 cell-derived PEDV PT52 S with a CTD-open conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Hsu, S.T.D, Draczkowski, P, Wang, Y.S. | | Deposit date: | 2022-06-21 | | Release date: | 2022-08-03 | | Last modified: | 2022-09-14 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | In situ structure and dynamics of an alphacoronavirus spike protein by cryo-ET and cryo-EM.
Nat Commun, 13, 2022
|
|
7Y6S
 
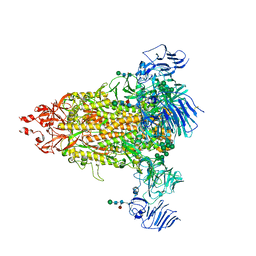 | | Cryo-EM map of IPEC-J2 cell-derived PEDV PT52 S protein with three D0-up | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Hsu, S.T.D, Draczkowski, P, Wang, Y.S. | | Deposit date: | 2022-06-21 | | Release date: | 2022-08-03 | | Last modified: | 2022-09-14 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | In situ structure and dynamics of an alphacoronavirus spike protein by cryo-ET and cryo-EM.
Nat Commun, 13, 2022
|
|
7Y6T
 
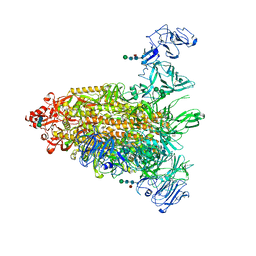 | | Cryo-EM map of IPEC-J2 cell-derived PEDV PT52 S protein one D0-down and two D0-up | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Hsu, S.T.D, Draczkowski, P, Wang, Y.S. | | Deposit date: | 2022-06-21 | | Release date: | 2022-08-03 | | Last modified: | 2023-07-26 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | In situ structure and dynamics of an alphacoronavirus spike protein by cryo-ET and cryo-EM.
Nat Commun, 13, 2022
|
|
2ZUB
 
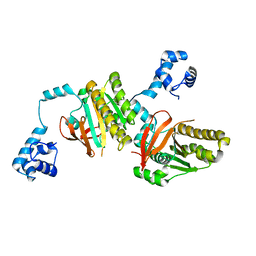 | | Left handed RadA | | Descriptor: | DNA repair and recombination protein radA | | Authors: | Chang, Y.W, Ko, T.P, Wang, T.F, Wang, A.H.J. | | Deposit date: | 2008-10-15 | | Release date: | 2009-04-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Three new structures of left-handed RADA helical filaments: structural flexibility of N-terminal domain is critical for recombinase activity
Plos One, 4, 2009
|
|
2ZUD
 
 | |
5ZME
 
 | |
5ZMF
 
 | | AMPPNP complex of C. reinhardtii ArsA1 | | Descriptor: | ATPase ARSA1, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Lin, T.W, Hsiao, C.D, Chang, H.Y. | | Deposit date: | 2018-04-03 | | Release date: | 2019-03-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.556 Å) | | Cite: | Structural analysis of chloroplast tail-anchored membrane protein recognition by ArsA1.
Plant J., 99, 2019
|
|
2ZUC
 
 | |
8KCM
 
 | | MmCPDII-DNA complex containing low-dosage, light induced repaired DNA. | | Descriptor: | Deoxyribodipyrimidine photo-lyase, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Maestre-Reyna, M, Wang, P.-H, Nango, E, Hosokawa, Y, Saft, M, Furrer, A, Yang, C.-H, Ngura Putu, E.P.G, Wu, W.-J, Emmerich, H.-J, Engilberge, S, Caramello, N, Wranik, M, Glover, H.L, Franz-Badur, S, Wu, H.-Y, Lee, C.-C, Huang, W.-C, Huang, K.-F, Chang, Y.-K, Liao, J.-H, Weng, J.-H, Gad, W, Chang, C.-W, Pang, A.H, Gashi, D, Beale, E, Ozerov, D, Milne, C, Cirelli, C, Bacellar, C, Sugahara, M, Owada, S, Joti, Y, Yamashita, A, Tanaka, R, Tanaka, T, Luo, F.J, Tono, K, Kiontke, S, Spadaccini, R, Royant, A, Yamamoto, J, Iwata, S, Standfuss, J, Essen, L.-O, Bessho, Y, Tsai, M.-D. | | Deposit date: | 2023-08-08 | | Release date: | 2023-11-22 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Visualizing the DNA repair process by a photolyase at atomic resolution.
Science, 382, 2023
|
|
2DFL
 
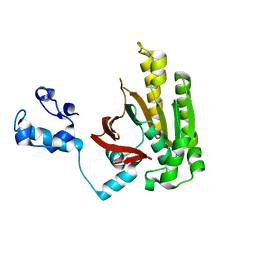 | | Crystal structure of left-handed RadA filament | | Descriptor: | DNA repair and recombination protein radA | | Authors: | Chen, L.T, Ko, T.P, Wang, T.F, Wang, A.H.J. | | Deposit date: | 2006-03-02 | | Release date: | 2007-01-23 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of the left-handed archaeal RadA helical filament: identification of a functional motif for controlling quaternary structures and enzymatic functions of RecA family proteins
Nucleic Acids Res., 35, 2007
|
|
6JC5
 
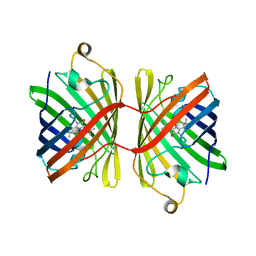 | |
6JC6
 
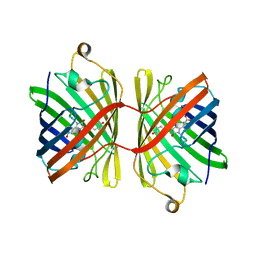 | |
7F62
 
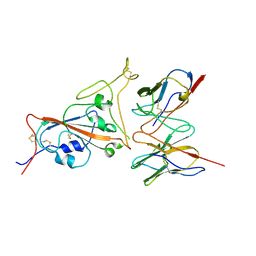 | | Cryo-EM structure of SARS-CoV-2 spike in complex with a neutralizing antibody chAb-25 (Focused refinement of S-RBD and chAb-25 region) | | Descriptor: | RBD-chAb-25, Heavy chain, Light chain, ... | | Authors: | Yang, T.J, Yu, P.Y, Wu, H.C, Hsu, S.T.D. | | Deposit date: | 2021-06-24 | | Release date: | 2021-08-04 | | Last modified: | 2022-02-16 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structure-guided antibody cocktail for prevention and treatment of COVID-19.
Plos Pathog., 17, 2021
|
|
7F63
 
 | | Cryo-EM structure of SARS-CoV-2 spike in complex with a neutralizing antibody chAb-45 (Focused refinement of S-RBD and chAb-45 region) | | Descriptor: | RBD-chAb45, Heavy chain, Light chain, ... | | Authors: | Yang, T.J, Yu, P.Y, Wu, H.C, Hsu, S.T.D. | | Deposit date: | 2021-06-24 | | Release date: | 2021-08-04 | | Last modified: | 2022-02-16 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structure-guided antibody cocktail for prevention and treatment of COVID-19.
Plos Pathog., 17, 2021
|
|
4EJT
 
 | |
8K1T
 
 | | Potassium transporter KtrAB from Bacillus subtilis in ATP-bound state with addition of MgCl2 | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Ktr system potassium uptake protein A, Ktr system potassium uptake protein B, ... | | Authors: | Chang, Y.K, Chiang, W.T, Hu, N.J, Tsai, M.D. | | Deposit date: | 2023-07-11 | | Release date: | 2024-04-03 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (2.48 Å) | | Cite: | Structural basis and synergism of ATP and Na+ activation in bacterial K+ uptake system KtrAB
Nat Commun, 15, 2024
|
|
8K1U
 
 | | Potassium transporter KtrAB from Bacillus subtilis in ATP-bound state with addition of EDTA and EGTA | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Ktr system potassium uptake protein A, Ktr system potassium uptake protein B, ... | | Authors: | Chang, Y.K, Chiang, W.T, Hu, N.J, Tsai, M.D. | | Deposit date: | 2023-07-11 | | Release date: | 2024-04-03 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (2.82 Å) | | Cite: | Structural basis and synergism of ATP and Na+ activation in bacterial K+ uptake system KtrAB
Nat Commun, 15, 2024
|
|
