5ENM
 
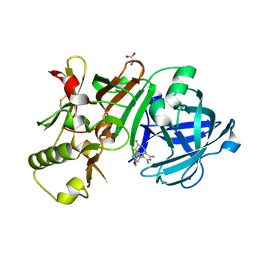 | | Compound 10 | | Descriptor: | (2~{R},4~{S},6~{S})-4-[2,4-bis(fluoranyl)-5-pyrimidin-5-yl-phenyl]-6-(3,5-dimethyl-1,2-oxazol-4-yl)-1,3-thiazinan-2-amine, Beta-secretase 1, GLYCEROL, ... | | Authors: | Lewis, H.A. | | Deposit date: | 2015-11-09 | | Release date: | 2016-10-05 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Targeting the BACE1 Active Site Flap Leads to a Potent Inhibitor That Elicits Robust Brain A beta Reduction in Rodents.
Acs Med.Chem.Lett., 7, 2016
|
|
5ENK
 
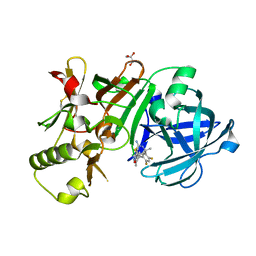 | | Compound 18 | | Descriptor: | (4~{S},6~{S})-4-[2,4-bis(fluoranyl)-5-pyrimidin-5-yl-phenyl]-6-(3,5-dimethyl-1,2-oxazol-4-yl)-4-methyl-5,6-dihydro-1,3-thiazin-2-amine, Beta-secretase 1, GLYCEROL, ... | | Authors: | Lewis, H.A. | | Deposit date: | 2015-11-09 | | Release date: | 2016-07-06 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Targeting the BACE1 Active Site Flap Leads to a Potent Inhibitor That Elicits Robust Brain A beta Reduction in Rodents.
Acs Med.Chem.Lett., 7, 2016
|
|
6AMN
 
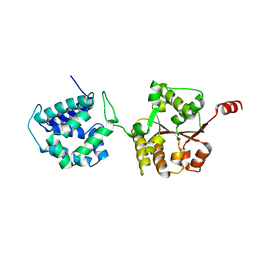 | | Crystal Structure of Hsp104 N Domain | | Descriptor: | Heat shock protein 104 | | Authors: | Lee, S. | | Deposit date: | 2017-08-10 | | Release date: | 2017-11-29 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.816 Å) | | Cite: | Overlapping and Specific Functions of the Hsp104 N Domain Define Its Role in Protein Disaggregation.
Sci Rep, 7, 2017
|
|
7V1R
 
 | | Leifsonia Alcohol Dehydrogenases LnADH | | Descriptor: | Alcohol Dehydrogenases, ISOPROPYL ALCOHOL, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Song, Y, Qu, X. | | Deposit date: | 2021-08-05 | | Release date: | 2021-12-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Engineering Leifsonia Alcohol Dehydrogenase for Thermostability and Catalytic Efficiency by Enhancing Subunit Interactions.
Chembiochem, 22, 2021
|
|
7V1Q
 
 | | Leifsonia Alcohol Dehydrogenases LnADH | | Descriptor: | Alcohol Dehydrogenases, ISOPROPYL ALCOHOL, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Song, Y, Qu, X. | | Deposit date: | 2021-08-05 | | Release date: | 2021-12-29 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Engineering Leifsonia Alcohol Dehydrogenase for Thermostability and Catalytic Efficiency by Enhancing Subunit Interactions.
Chembiochem, 22, 2021
|
|
5BPY
 
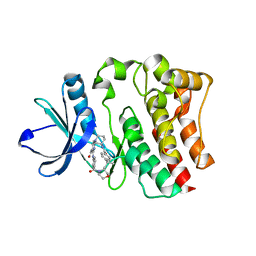 | |
7WTE
 
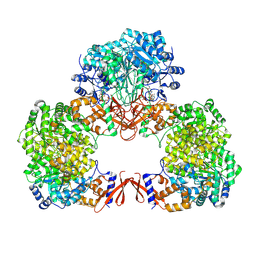 | | Cryo-EM structure of human pyruvate carboxylase with acetyl-CoA in the intermediate state 2 | | Descriptor: | ACETYL COENZYME *A, ADENOSINE-5'-TRIPHOSPHATE, Pyruvate carboxylase, ... | | Authors: | Chai, P, Lan, P, Wu, J, Lei, M. | | Deposit date: | 2022-02-04 | | Release date: | 2022-11-09 | | Last modified: | 2022-11-16 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Mechanistic insight into allosteric activation of human pyruvate carboxylase by acetyl-CoA.
Mol.Cell, 82, 2022
|
|
7WTB
 
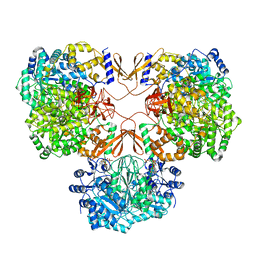 | | Cryo-EM structure of human pyruvate carboxylase with acetyl-CoA | | Descriptor: | 5-(HEXAHYDRO-2-OXO-1H-THIENO[3,4-D]IMIDAZOL-6-YL)PENTANAL, ACETYL COENZYME *A, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Chai, P, Lan, P, Wu, J, Lei, M. | | Deposit date: | 2022-02-04 | | Release date: | 2022-11-09 | | Last modified: | 2022-11-16 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Mechanistic insight into allosteric activation of human pyruvate carboxylase by acetyl-CoA.
Mol.Cell, 82, 2022
|
|
7WTA
 
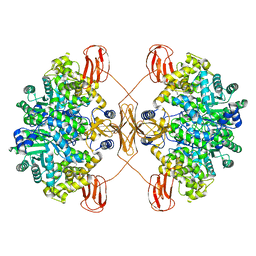 | | Cryo-EM structure of human pyruvate carboxylase in apo state | | Descriptor: | 5-(HEXAHYDRO-2-OXO-1H-THIENO[3,4-D]IMIDAZOL-6-YL)PENTANAL, Pyruvate carboxylase, mitochondrial | | Authors: | Chai, P, Lan, P, Wu, J, Lei, M. | | Deposit date: | 2022-02-04 | | Release date: | 2022-11-09 | | Last modified: | 2022-11-16 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Mechanistic insight into allosteric activation of human pyruvate carboxylase by acetyl-CoA.
Mol.Cell, 82, 2022
|
|
7WTD
 
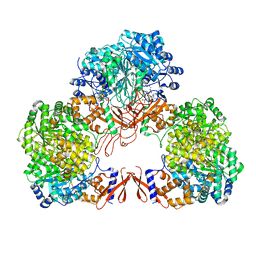 | | Cryo-EM structure of human pyruvate carboxylase with acetyl-CoA in the intermediate state 1 | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, COENZYME A, Pyruvate carboxylase, ... | | Authors: | Chai, P, Lan, P, Wu, J, Lei, M. | | Deposit date: | 2022-02-04 | | Release date: | 2022-11-09 | | Last modified: | 2022-11-16 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Mechanistic insight into allosteric activation of human pyruvate carboxylase by acetyl-CoA.
Mol.Cell, 82, 2022
|
|
7WTC
 
 | | Cryo-EM structure of human pyruvate carboxylase with acetyl-CoA in the ground state | | Descriptor: | 5-(HEXAHYDRO-2-OXO-1H-THIENO[3,4-D]IMIDAZOL-6-YL)PENTANAL, ACETYL COENZYME *A, Pyruvate carboxylase, ... | | Authors: | Chai, P, Lan, P, Wu, J, Lei, M. | | Deposit date: | 2022-02-04 | | Release date: | 2022-11-09 | | Last modified: | 2022-11-16 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Mechanistic insight into allosteric activation of human pyruvate carboxylase by acetyl-CoA.
Mol.Cell, 82, 2022
|
|
5BQ0
 
 | |
5TOL
 
 | | CRYSTAL STRUCTURE OF BETA-SITE APP-CLEAVING ENZYME 1 COMPLEXED WITH N-(3-((4AS,7AS)-2-AMINO-4,4A,5,6-TETRAHYDRO-7AH-FURO[2,3-D][1,3]THIAZIN-7A-YL)-4-FLUOROPHENYL)-5-BROMO-2-PYRIDINECARBOXAMIDE | | Descriptor: | Beta-secretase 1, N-{3-[(4aR,7aR)-2-amino-4,4a,5,6-tetrahydro-7aH-furo[2,3-d][1,3]thiazin-7a-yl]-4-fluorophenyl}-5-bromopyridine-2-carboxamide | | Authors: | Muckelbauer, J.K. | | Deposit date: | 2016-10-18 | | Release date: | 2016-11-23 | | Last modified: | 2016-12-07 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Discovery of furo[2,3-d][1,3]thiazinamines as beta amyloid cleaving enzyme-1 (BACE1) inhibitors.
Bioorg.Med.Chem.Lett., 26, 2016
|
|
3NW7
 
 | | Crystal structure of insulin-like growth factor 1 receptor (IGF-1R-WT) complex with a carbon-linked proline isostere inhibitor (34) | | Descriptor: | Insulin-like growth factor 1 receptor, N-({4-[(3-cyclopropyl-1H-pyrazol-5-yl)amino]pyrrolo[2,1-f][1,2,4]triazin-2-yl}methyl)-6-fluoropyridine-3-carboxamide | | Authors: | Sack, J.S. | | Deposit date: | 2010-07-09 | | Release date: | 2010-07-28 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Proline isosteres in a series of 2,4-disubstituted pyrrolo[1,2-f][1,2,4]triazine inhibitors of IGF-1R kinase and IR kinase.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
4I5C
 
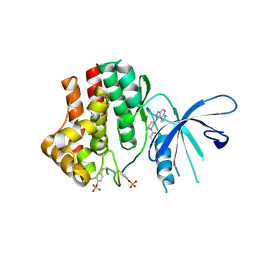 | | The Jak1 kinase domain in complex with inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 3-oxo-3-[(3R)-3-(pyrrolo[2,3-b][1,2,3]triazolo[4,5-d]pyridin-1(6H)-yl)piperidin-1-yl]propanenitrile, Tyrosine-protein kinase JAK1 | | Authors: | Fong, R, Lupardus, P.J. | | Deposit date: | 2012-11-28 | | Release date: | 2013-05-22 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Novel triazolo-pyrrolopyridines as inhibitors of Janus kinase 1.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
1FK2
 
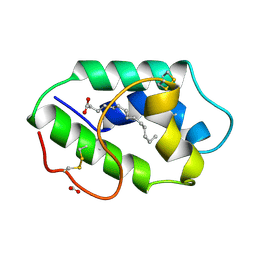 | | STRUCTURAL BASIS OF NON-SPECIFIC LIPID BINDING IN MAIZE LIPID-TRANSFER PROTEIN COMPLEXES WITH MYRISTIC ACID REVEALED BY HIGH-RESOLUTION X-RAY CRYSTALLOGRAPHY | | Descriptor: | FORMIC ACID, MYRISTIC ACID, NONSPECIFIC LIPID-TRANSFER PROTEIN | | Authors: | Han, G.W, Lee, J.Y, Song, H.K, Shin, D.H, Suh, S.W. | | Deposit date: | 2000-08-09 | | Release date: | 2001-06-06 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis of non-specific lipid binding in maize lipid-transfer protein complexes revealed by high-resolution X-ray crystallography.
J.Mol.Biol., 308, 2001
|
|
1FK7
 
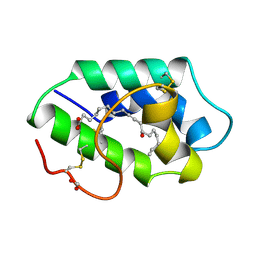 | | STRUCTURAL BASIS OF NON-SPECIFIC LIPID BINDING IN MAIZE LIPID-TRANSFER PROTEIN COMPLEXES WITH RICINOLEIC ACID REVEALED BY HIGH-RESOLUTION X-RAY CRYSTALLOGRAPHY | | Descriptor: | FORMIC ACID, NON-SPECIFIC LIPID TRANSFER PROTEIN, RICINOLEIC ACID | | Authors: | Han, G.W, Lee, J.Y, Song, H.K, Shin, D.H, Suh, S.W. | | Deposit date: | 2000-08-09 | | Release date: | 2001-06-06 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis of non-specific lipid binding in maize lipid-transfer protein complexes revealed by high-resolution X-ray crystallography.
J.Mol.Biol., 308, 2001
|
|
4I1D
 
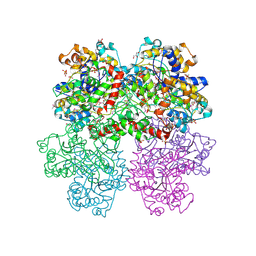 | | The crystal structure of an ABC transporter substrate-binding protein from Bradyrhizobium japonicum USDA 110 | | Descriptor: | ABC transporter substrate-binding protein, ACETATE ION, D-MALATE, ... | | Authors: | Fan, Y, Tan, K, Mack, J, Zerbs, S, Collart, F, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2012-11-20 | | Release date: | 2012-12-05 | | Last modified: | 2013-09-25 | | Method: | X-RAY DIFFRACTION (2.201 Å) | | Cite: | Structural and functional characterization of solute binding proteins for aromatic compounds derived from lignin: p-Coumaric acid and related aromatic acids.
Proteins, 81, 2013
|
|
1FK3
 
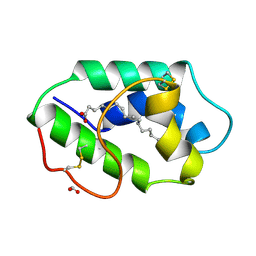 | | STRUCTURAL BASIS OF NON-SPECIFIC LIPID BINDING IN MAIZE LIPID-TRANSFER PROTEIN COMPLEXES WITH PALMITOLEIC ACID REVEALED BY HIGH-RESOLUTION X-RAY CRYSTALLOGRAPHY | | Descriptor: | FORMIC ACID, NONSPECIFIC LIPID-TRANSFER PROTEIN, PALMITOLEIC ACID | | Authors: | Han, G.W, Lee, J.Y, Song, H.K, Shin, D.H, Suh, S.W. | | Deposit date: | 2000-08-09 | | Release date: | 2001-06-06 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis of non-specific lipid binding in maize lipid-transfer protein complexes revealed by high-resolution X-ray crystallography.
J.Mol.Biol., 308, 2001
|
|
1FK0
 
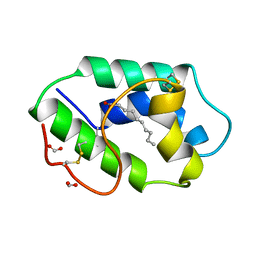 | | STRUCTURAL BASIS OF NON-SPECIFIC LIPID BINDING IN MAIZE LIPID-TRANSFER PROTEIN COMPLEXES WITH CAPRIC ACID REVEALED BY HIGH-RESOLUTION X-RAY CRYSTALLOGRAPHY | | Descriptor: | DECANOIC ACID, FORMIC ACID, NONSPECIFIC LIPID-TRANSFER PROTEIN | | Authors: | Han, G.W, Lee, J.Y, Song, H.K, Shin, D.H, Suh, S.W. | | Deposit date: | 2000-08-08 | | Release date: | 2001-06-06 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis of non-specific lipid binding in maize lipid-transfer protein complexes revealed by high-resolution X-ray crystallography.
J.Mol.Biol., 308, 2001
|
|
1FK5
 
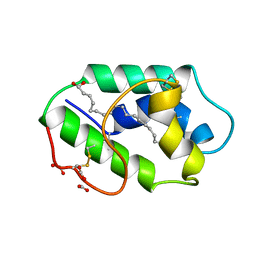 | | STRUCTURAL BASIS OF NON-SPECIFIC LIPID BINDING IN MAIZE LIPID-TRANSFER PROTEIN COMPLEXES WITH OLEIC ACID REVEALED BY HIGH-RESOLUTION X-RAY CRYSTALLOGRAPHY | | Descriptor: | FORMIC ACID, NONSPECIFIC LIPID-TRANSFER PROTEIN, OLEIC ACID | | Authors: | Han, G.W, Lee, J.Y, Song, H.K, Shin, D.H, Suh, S.W. | | Deposit date: | 2000-08-09 | | Release date: | 2001-06-06 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structural basis of non-specific lipid binding in maize lipid-transfer protein complexes revealed by high-resolution X-ray crystallography.
J.Mol.Biol., 308, 2001
|
|
1FK4
 
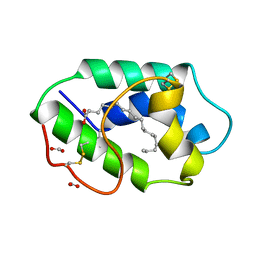 | | STRUCTURAL BASIS OF NON-SPECIFIC LIPID BINDING IN MAIZE LIPID-TRANSFER PROTEIN COMPLEXES WITH STEARIC ACID REVEALED BY HIGH-RESOLUTION X-RAY CRYSTALLOGRAPHY | | Descriptor: | FORMIC ACID, NONSPECIFIC LIPID-TRANSFER PROTEIN, STEARIC ACID | | Authors: | Han, G.W, Lee, J.Y, Song, H.K, Shin, D.H, Suh, S.W. | | Deposit date: | 2000-08-09 | | Release date: | 2001-06-06 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis of non-specific lipid binding in maize lipid-transfer protein complexes revealed by high-resolution X-ray crystallography.
J.Mol.Biol., 308, 2001
|
|
1FK1
 
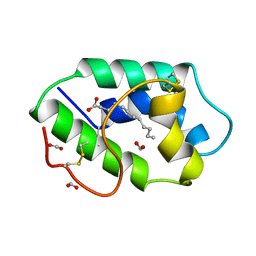 | | STRUCTURAL BASIS OF NON-SPECIFIC LIPID BINDING IN MAIZE LIPID-TRANSFER PROTEIN COMPLEXES WITH LAURIC ACID REVEALED BY HIGH-RESOLUTION X-RAY CRYSTALLOGRAPHY | | Descriptor: | FORMIC ACID, LAURIC ACID, NON-SPECIFIC LIPID TRANSFER PROTEIN | | Authors: | Han, G.W, Lee, J.Y, Song, H.K, Shin, D.H, Suh, S.W. | | Deposit date: | 2000-08-09 | | Release date: | 2001-06-06 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis of non-specific lipid binding in maize lipid-transfer protein complexes revealed by high-resolution X-ray crystallography.
J.Mol.Biol., 308, 2001
|
|
1FK6
 
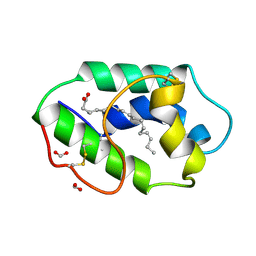 | | STRUCTURAL BASIS OF NON-SPECIFIC LIPID BINDING IN MAIZE LIPID-TRANSFER PROTEIN COMPLEXES WITH ALPHA-LINOLENIC ACID REVEALED BY HIGH-RESOLUTION X-RAY CRYSTALLOGRAPHY | | Descriptor: | ALPHA-LINOLENIC ACID, FORMIC ACID, NON-SPECIFIC LIPID TRANSFER PROTEIN | | Authors: | Han, G.W, Lee, J.Y, Song, H.K, Shin, D.H, Suh, S.W. | | Deposit date: | 2000-08-09 | | Release date: | 2001-06-06 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis of non-specific lipid binding in maize lipid-transfer protein complexes revealed by high-resolution X-ray crystallography.
J.Mol.Biol., 308, 2001
|
|
1FA2
 
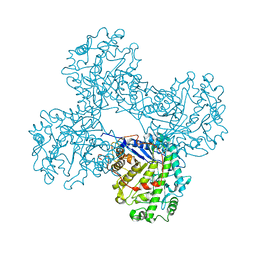 | | CRYSTAL STRUCTURE OF BETA-AMYLASE FROM SWEET POTATO | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, BETA-AMYLASE, alpha-D-glucopyranose-(1-4)-2-deoxy-beta-D-arabino-hexopyranose | | Authors: | Lee, B.I, Cheong, C.G, Suh, S.W. | | Deposit date: | 2000-07-12 | | Release date: | 2000-08-16 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystallization, molecular replacement solution, and refinement of tetrameric beta-amylase from sweet potato.
Proteins, 21, 1995
|
|
