2PZH
 
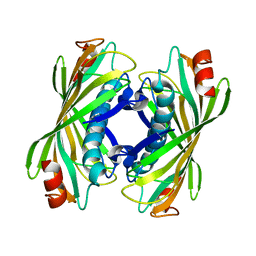 | | YbgC thioesterase (Hp0496) from Helicobacter pylori | | Descriptor: | Hypothetical protein HP_0496 | | Authors: | Angelini, A, Cendron, L, Goncalves, S, Zanotti, G, Terradot, L. | | Deposit date: | 2007-05-18 | | Release date: | 2008-04-08 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural and enzymatic characterization of HP0496, a YbgC thioesterase from Helicobacter pylori.
Proteins, 72, 2008
|
|
3HPE
 
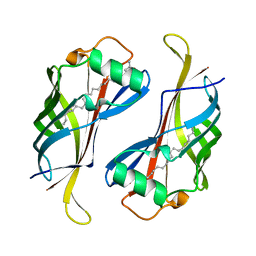 | |
7ZRT
 
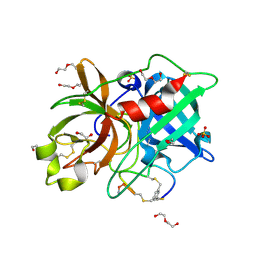 | | Crystal structure of human Urokinase-type plasminogen activator in complex with bicycle peptide inhibitor UK970 | | Descriptor: | 1,2-ETHANEDIOL, 1,3,5-tris(bromomethyl)benzene, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Caregnato, A, Angela, P, Mazzoccato, Y, Frasson, N, Angelini, A, Cendron, L. | | Deposit date: | 2022-05-05 | | Release date: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of human Urokinase-type plasminogen activator in complex with bicycle peptide inhibitor UK970
To Be Published
|
|
7ZRR
 
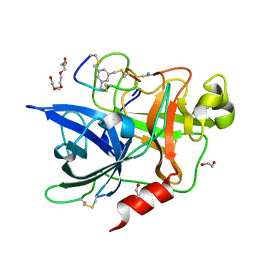 | | Crystal structure of human Urokinase-type plasminogen activator in complex with bicycle peptide inhibitor UK965 | | Descriptor: | 1,2-ETHANEDIOL, 1,3,5-tris(bromomethyl)benzene, AMINO GROUP, ... | | Authors: | Caregnato, A, Angela, P, Mazzoccato, Y, Frasson, N, Angelini, A, Cendron, L. | | Deposit date: | 2022-05-05 | | Release date: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Crystal structure of human Urokinase-type plasminogen activator in complex with bicycle peptide inhibitor UK965
To Be Published
|
|
4KZS
 
 | | Crystal structure of the secreted protein HP1454 from the human pathogen Helicobacter pylori | | Descriptor: | LPP20 lipofamily protein, PENTAETHYLENE GLYCOL | | Authors: | Quarantini, S, Cendron, L, Fischer, W, Zanotti, G. | | Deposit date: | 2013-05-30 | | Release date: | 2014-06-04 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of the secreted protein HP1454 from the human pathogen Helicobacter pylori.
Proteins, 82, 2014
|
|
4LS3
 
 | | THE crystal STRUCTURE OF HELICOBACTER PYLORI CEUE(HP1561)/NI-HIS COMPL | | Descriptor: | HISTIDINE, NICKEL (II) ION, Nickel (III) ABC transporter, ... | | Authors: | Salamina, M, Shaik, M.M, Cendron, L, Zanotti, G. | | Deposit date: | 2013-07-22 | | Release date: | 2014-01-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Helicobacter pylori periplasmic receptor CeuE (HP1561) modulates its nickel affinity via organic metallophores.
Mol.Microbiol., 91, 2014
|
|
3SXP
 
 | |
7BN6
 
 | | Crystal structure of G. sulphuraria ene-reductase GsOYE in complex with b-angelica lactone | | Descriptor: | (2~{R})-2-methyl-2~{H}-furan-5-one, FLAVIN MONONUCLEOTIDE, NADPH2 dehydrogenase-like protein | | Authors: | Robescu, M.S, Bergantino, E, Hall, M, Cendron, L. | | Deposit date: | 2021-01-21 | | Release date: | 2022-07-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Asymmetric Proton Transfer Catalysis by Stereocomplementary Old Yellow Enzymes for C=C Bond Isomerization Reaction
Acs Catalysis, 12, 2022
|
|
7BN7
 
 | | Crystal structure of ene-reductase OYE2 from S. cerevisiae | | Descriptor: | FLAVIN MONONUCLEOTIDE, GLYCEROL, OYE2 isoform 1 | | Authors: | Robescu, M.S, Bergantino, E, Hall, M, Cendron, L. | | Deposit date: | 2021-01-21 | | Release date: | 2022-07-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Asymmetric Proton Transfer Catalysis by Stereocomplementary Old Yellow Enzymes for C=C Bond Isomerization Reaction
Acs Catalysis, 12, 2022
|
|
7BO0
 
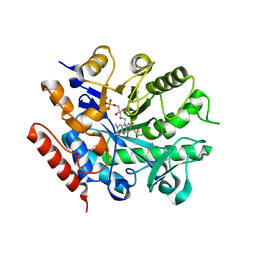 | | Crystal structure of ene-reductase GsOYE from Galdieria sulphuraria in complex with alpha-angelica lactone | | Descriptor: | 5-methyl-3~{H}-furan-2-one, FLAVIN MONONUCLEOTIDE, NADPH2 dehydrogenase-like protein | | Authors: | Robescu, M.S, Bergantino, E, Hall, M, Cendron, L. | | Deposit date: | 2021-01-23 | | Release date: | 2022-07-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Asymmetric Proton Transfer Catalysis by Stereocomplementary Old Yellow Enzymes for C=C Bond Isomerization Reaction
Acs Catalysis, 12, 2022
|
|
7BLF
 
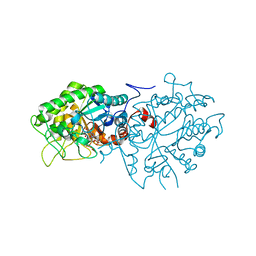 | | Crystal structure of ene-reductase OYE4 from Botryotinia fuckeliana (BfOYE4) | | Descriptor: | FLAVIN MONONUCLEOTIDE, FORMIC ACID, Oxidored_FMN domain-containing protein | | Authors: | Robescu, M.S, Bergantino, E, Hall, M, Cendron, L. | | Deposit date: | 2021-01-18 | | Release date: | 2022-07-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Asymmetric Proton Transfer Catalysis by Stereocomplementary Old Yellow Enzymes for C=C Bond Isomerization Reaction
Acs Catalysis, 12, 2022
|
|
3TLM
 
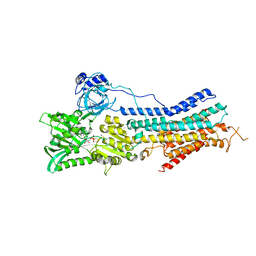 | | Crystal Structure of Endoplasmic Reticulum Ca2+-ATPase (SERCA) From Bovine Muscle | | Descriptor: | CALCIUM ION, MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, ... | | Authors: | Sacchetto, R, Bertipaglia, I, Giannetti, S, Cendron, L, Mascarello, F, Damiani, E, Carafoli, E, Zanotti, G. | | Deposit date: | 2011-08-30 | | Release date: | 2012-03-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Crystal structure of sarcoplasmic reticulum Ca(2+)-ATPase (SERCA) from bovine muscle.
J.Struct.Biol., 178, 2012
|
|
6TGD
 
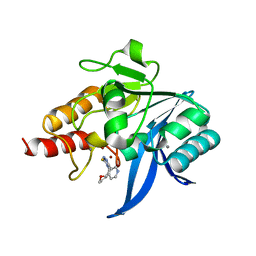 | | Crystal structure of NDM-1 in complex with triazole-based inhibitor OP31 | | Descriptor: | 4-[[(2~{S})-oxolan-2-yl]methyl]-3-pyridin-3-yl-1~{H}-1,2,4-triazole-5-thione, CALCIUM ION, Metallo-beta-lactamase NDM-1, ... | | Authors: | Maso, L, Spirakis, F, Santucci, M, Simon, C, Docquier, J.D, Cruciani, G, Costi, M.P, Tondi, D, Cendron, L. | | Deposit date: | 2019-11-15 | | Release date: | 2020-08-26 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | Virtual screening identifies broad-spectrum beta-lactamase inhibitors with activity on clinically relevant serine- and metallo-carbapenemases.
Sci Rep, 10, 2020
|
|
6TGI
 
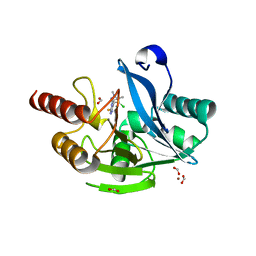 | | Crystal structure of VIM-2 in complex with triazole-based inhibitor OP24 | | Descriptor: | 5-(4-chloranyl-1,5-dimethyl-pyrazol-3-yl)-4-ethyl-1,2,4-triazole-3-thiol, FORMIC ACID, Vim-1, ... | | Authors: | Maso, L, Spirakis, F, Santucci, M, Simon, C, Docquier, J.D, Cruciani, G, Costi, M.P, Tondi, D, Cendron, L. | | Deposit date: | 2019-11-15 | | Release date: | 2020-10-14 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Virtual screening identifies broad-spectrum beta-lactamase inhibitors with activity on clinically relevant serine- and metallo-carbapenemases.
Sci Rep, 10, 2020
|
|
6TS9
 
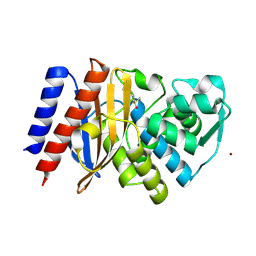 | | Crystal structure of GES-5 carbapenemase | | Descriptor: | 1,2-ETHANEDIOL, BROMIDE ION, Beta-lactamase, ... | | Authors: | Maso, L, Tondi, D, Klein, R, Montanari, M, Bellio, C, Celenza, G, Brenk, R, Cendron, L. | | Deposit date: | 2019-12-20 | | Release date: | 2020-03-04 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Targeting the Class A Carbapenemase GES-5 via Virtual Screening.
Biomolecules, 10, 2020
|
|
4INP
 
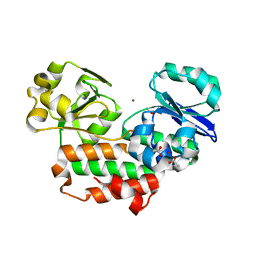 | | The crystal structure of Helicobacter pylori Ceue (HP1561) with Ni(II) bound | | Descriptor: | ACETATE ION, Iron (III) ABC transporter, periplasmic iron-binding protein, ... | | Authors: | Shaik, M.M, Cendron, L, Zanotti, G. | | Deposit date: | 2013-01-05 | | Release date: | 2014-01-08 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Helicobacter pylori periplasmic receptor CeuE (HP1561) modulates its nickel affinity via organic metallophores.
Mol.Microbiol., 91, 2014
|
|
4INO
 
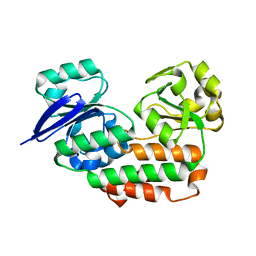 | |
4INN
 
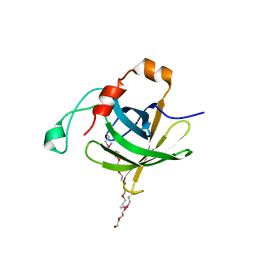 | | Protein HP1028 from the human pathogen Helicobacter pylori belongs to the lipocalin family | | Descriptor: | 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, HP1028 | | Authors: | Barison, N, Cendron, L, Loconte, V, Zanotti, G. | | Deposit date: | 2013-01-05 | | Release date: | 2013-07-31 | | Last modified: | 2014-02-05 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Protein HP1028 from the human pathogen Helicobacter pylori belongs to the lipocalin family.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
3BT0
 
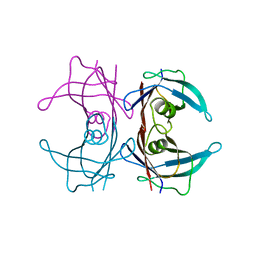 | | Crystal structure of transthyretin variant V20S | | Descriptor: | Transthyretin | | Authors: | Zanotti, G, Folli, C, Cendron, L, Gliubich, F, Negro, A, Berni, R. | | Deposit date: | 2007-12-27 | | Release date: | 2008-11-11 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Structural and mutational analyses of protein-protein interactions between transthyretin and retinol-binding protein.
Febs J., 275, 2008
|
|
2H1X
 
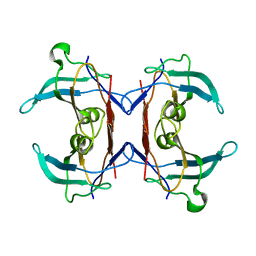 | | Crystal structure of 5-hydroxyisourate Hydrolase (formerly known as TRP, Transthyretin Related Protein) | | Descriptor: | 5-hydroxyisourate Hydrolase (formerly known as TRP, Transthyretin Related Protein) | | Authors: | Zanotti, G, Cendron, L, Folli, C, Ramazzina, I, Percudani, R, Berni, R. | | Deposit date: | 2006-05-17 | | Release date: | 2006-10-31 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structure of Zebra fish HIUase: Insights into Evolution of an Enzyme to a Hormone Transporter.
J.Mol.Biol., 363, 2006
|
|
2H6U
 
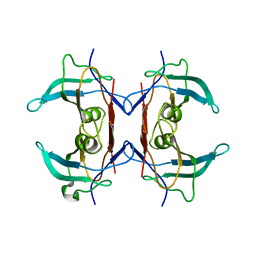 | | Crystal structure of 5-hydroxyisourate hydrolase (formerly known as TRP, transthyretin related protein) | | Descriptor: | 5-HYDROXYISOURATE HYDROLASE (FORMERLY KNOWN AS TRP, TRANSTHYRETIN RELATED PROTEIN) | | Authors: | Zanotti, G, Cendron, L, Folli, C, Ramazzina, I, Percudani, R, Berni, R. | | Deposit date: | 2006-06-01 | | Release date: | 2006-10-31 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of Zebra fish HIUase: Insights into Evolution of an Enzyme to a Hormone Transporter.
J.Mol.Biol., 363, 2006
|
|
3CEI
 
 | | Crystal Structure of Superoxide Dismutase from Helicobacter pylori | | Descriptor: | FE (III) ION, SULFATE ION, Superoxide dismutase | | Authors: | Esposito, L, Seydel, A, Aiello, R, Sorrentino, G, Cendron, L, Zanotti, G, Zagari, A. | | Deposit date: | 2008-02-29 | | Release date: | 2008-06-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The crystal structure of the superoxide dismutase from Helicobacter pylori reveals a structured C-terminal extension
Biochim.Biophys.Acta, 1784, 2008
|
|
2NOY
 
 | | Crystal structure of transthyretin mutant I84S at PH 7.5 | | Descriptor: | Transthyretin | | Authors: | Pasquato, N, Berni, R, Folli, C, Alfieri, B, Cendron, L, Zanotti, G. | | Deposit date: | 2006-10-26 | | Release date: | 2007-01-16 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Acidic pH-induced conformational changes in amyloidogenic mutant transthyretin
J.Mol.Biol., 366, 2007
|
|
3IBX
 
 | | Crystal structure of F47Y variant of TenA (HP1287) from Helicobacter pylori | | Descriptor: | Putative thiaminase II | | Authors: | Barison, N, Cendron, L, Trento, A, Angelini, A, Zanotti, G. | | Deposit date: | 2009-07-17 | | Release date: | 2009-11-17 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural and mutational analysis of TenA protein (HP1287) from the Helicobacter pylori thiamin salvage pathway - evidence of a different substrate specificity.
Febs J., 276, 2009
|
|
3ISL
 
 | | Crystal structure of ureidoglycine-glyoxylate aminotransferase (pucG) from Bacillus subtilis | | Descriptor: | PYRIDOXAL-5'-PHOSPHATE, Purine catabolism protein pucG | | Authors: | Costa, R, Cendron, L, Ramazzina, I, Berni, R, Peracchi, A, Percudani, R, Zanotti, G. | | Deposit date: | 2009-08-26 | | Release date: | 2010-09-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Amino acids from purines in GUT bacteria
To be Published
|
|
