4CP5
 
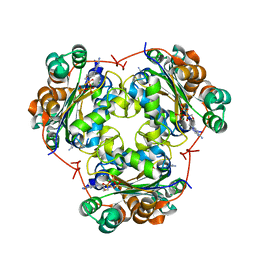 | | ndpK in complex with (Rp)-SPMPApp | | Descriptor: | NUCLEOSIDE DIPHOSPHATE KINASE, CYTOSOLIC, [[(2R)-1-(6-aminopurin-9-yl)propan-2-yl]oxymethyl-sulfanyl-phosphoryl] phosphono hydrogen phosphate | | Authors: | Priet, S, Ferron, F, Alvarez, K, Verron, M, Canard, B. | | Deposit date: | 2014-01-31 | | Release date: | 2015-02-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Enzymatic Synthesis of Acyclic Nucleoside Thiophosphonate Diphosphates: Effect of the Alpha-Phosphorus Configuration on HIV-1 RT Activity.
Antiviral Res., 117, 2015
|
|
2XYV
 
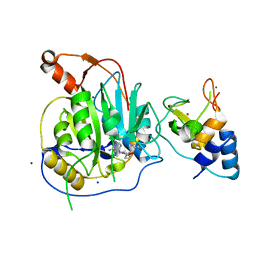 | | Crystal structure of the nsp16 nsp10 SARS coronavirus complex | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, NON-STRUCTURAL PROTEIN 10, ... | | Authors: | Decroly, E, Debarnot, C, Ferron, F, Bouvet, M, Coutard, B, Imbert, I, Gluais, L, Papageorgiou, N, Ortiz-Lombardia, M, Lescar, J, Canard, B. | | Deposit date: | 2010-11-19 | | Release date: | 2011-10-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Crystal Structure and Functional Analysis of the Sars-Coronavirus RNA CAP 2'-O-Methyltransferase Nsp10/Nsp16 Complex.
Plos Pathog., 7, 2011
|
|
2XYQ
 
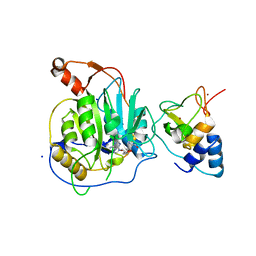 | | Crystal structure of the nsp16 nsp10 SARS coronavirus complex | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, NON-STRUCTURAL PROTEIN 10, ... | | Authors: | Decroly, E, Debarnot, C, Ferron, F, Bouvet, M, Coutard, B, Imbert, I, Gluais, L, Papageorgiou, N, Ortiz-Lombardia, M, Lescar, J, Canard, B. | | Deposit date: | 2010-11-18 | | Release date: | 2011-10-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure and Functional Analysis of the Sars-Coronavirus RNA CAP 2'-O-Methyltransferase Nsp10/Nsp16 Complex.
Plos Pathog., 7, 2011
|
|
2XYR
 
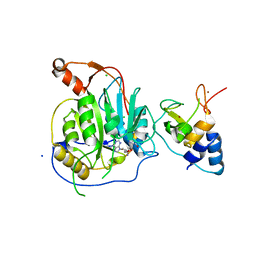 | | Crystal structure of the nsp16 nsp10 SARS coronavirus complex | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, NON-STRUCTURAL PROTEIN 10, ... | | Authors: | Decroly, E, Debarnot, C, Ferron, F, Bouvet, M, Coutard, B, Imbert, I, Gluais, L, Papageorgiou, N, Ortiz-Lombardia, M, Lescar, J, Canard, B. | | Deposit date: | 2010-11-18 | | Release date: | 2011-10-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure and Functional Analysis of the Sars-Coronavirus RNA CAP 2'-O-Methyltransferase Nsp10/Nsp16 Complex.
Plos Pathog., 7, 2011
|
|
7Z05
 
 | | White Bream virus N7-Methyltransferase | | Descriptor: | DI(HYDROXYETHYL)ETHER, Non-structural protein 1, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Shannon, A, Gauffre, P, Canard, B, Ferron, F. | | Deposit date: | 2022-02-22 | | Release date: | 2022-09-28 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | A second type of N7-guanine RNA cap methyltransferase in an unusual locus of a large RNA virus genome.
Nucleic Acids Res., 50, 2022
|
|
7Z2J
 
 | | White Bream virus N7-Methyltransferase | | Descriptor: | Non-structural protein 1, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Shannon, A, Gauffre, P, Canard, B, Ferron, F. | | Deposit date: | 2022-02-28 | | Release date: | 2022-11-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.657 Å) | | Cite: | A second type of N7-guanine RNA cap methyltransferase in an unusual locus of a large RNA virus genome.
Nucleic Acids Res., 50, 2022
|
|
3Q3X
 
 | | Crystal structure of the main protease (3C) from human enterovirus B EV93 | | Descriptor: | GLYCEROL, HEVB EV93 3C protease, MAGNESIUM ION | | Authors: | Costenaro, L, Sola, M, Coutard, B, Norder, H, Canard, B, Coll, M. | | Deposit date: | 2010-12-22 | | Release date: | 2011-09-07 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Basis for Antiviral Inhibition of the Main Protease, 3C, from Human Enterovirus 93.
J.Virol., 85, 2011
|
|
3Q3Y
 
 | | Complex structure of HEVB EV93 main protease 3C with Compound 1 (AG7404) | | Descriptor: | 1,2-ETHANEDIOL, AMMONIUM ION, HEVB EV93 3C protease, ... | | Authors: | Costenaro, L, Kaczmarska, Z, Arnan, C, Sola, M, Coutard, B, Norder, H, Canard, B, Coll, M. | | Deposit date: | 2010-12-22 | | Release date: | 2011-09-07 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | Structural Basis for Antiviral Inhibition of the Main Protease, 3C, from Human Enterovirus 93.
J.Virol., 85, 2011
|
|
2V6J
 
 | | Kokobera Virus Helicase: Mutant Met47Thr | | Descriptor: | RNA HELICASE | | Authors: | Speroni, S, De Colibus, L, Coutard, B, Canard, B, Mattevi, A. | | Deposit date: | 2007-07-18 | | Release date: | 2008-03-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure and Biochemical Analysis of Kokobera Virus Helicase
Proteins: Struct., Funct., Bioinf., 70, 2008
|
|
2V6I
 
 | | Kokobera Virus Helicase | | Descriptor: | PYROPHOSPHATE 2-, RNA HELICASE | | Authors: | Speroni, S, De Colibus, L, Coutard, B, Canard, B, Mattevi, A. | | Deposit date: | 2007-07-18 | | Release date: | 2008-03-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure and Biochemical Analysis of Kokobera Virus Helicase
Proteins: Struct., Funct., Bioinf., 70, 2008
|
|
3RUO
 
 | | Complex structure of HevB EV93 main protease 3C with Rupintrivir (AG7088) | | Descriptor: | 4-{2-(4-FLUORO-BENZYL)-6-METHYL-5-[(5-METHYL-ISOXAZOLE-3-CARBONYL)-AMINO]-4-OXO-HEPTANOYLAMINO}-5-(2-OXO-PYRROLIDIN-3-YL)-PENTANOIC ACID ETHYL ESTER, CHLORIDE ION, HEVB EV93 3C PROTEASE, ... | | Authors: | Kaczmarska, Z, Janowski, R, Costenaro, L, Coutard, B, Norder, H, Canard, B, Coll, M. | | Deposit date: | 2011-05-05 | | Release date: | 2011-09-07 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural Basis for Antiviral Inhibition of the Main Protease, 3C, from Human Enterovirus 93.
J.Virol., 85, 2011
|
|
2W5E
 
 | | Structural and biochemical analysis of human pathogenic astrovirus serine protease at 2.0 Angstrom resolution | | Descriptor: | CADMIUM ION, CHLORIDE ION, PUTATIVE SERINE PROTEASE | | Authors: | Speroni, S, Rohayem, J, Nenci, S, Bonivento, D, Robel, I, Barthel, J, Coutard, B, Canard, B, Mattevi, A. | | Deposit date: | 2008-12-10 | | Release date: | 2009-03-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and Biochemical Analysis of Human Pathogenic Astrovirus Serine Protease at 2.0 A Resolution.
J.Mol.Biol., 387, 2009
|
|
5NFY
 
 | | SARS-CoV nsp10/nsp14 dynamic complex | | Descriptor: | DI(HYDROXYETHYL)ETHER, Polyprotein 1ab, ZINC ION | | Authors: | Ferron, F, Gluais, L, Vonrhein, C, Bricogne, G, Canard, B, Imbert, I. | | Deposit date: | 2017-03-16 | | Release date: | 2018-01-10 | | Last modified: | 2019-02-20 | | Method: | X-RAY DIFFRACTION (3.382 Å) | | Cite: | Structural and molecular basis of mismatch correction and ribavirin excision from coronavirus RNA.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
4TU0
 
 | | CRYSTAL STRUCTURE OF CHIKUNGUNYA VIRUS NSP3 MACRO DOMAIN IN COMPLEX WITH A 2'-5' OLIGOADENYLATE TRIMER | | Descriptor: | 2'-5' OLIGOADENYLATE TRIMER, DI(HYDROXYETHYL)ETHER, Non-structural polyprotein 3 | | Authors: | Morin, B, Ferron, f.p, Malet, h, Coutard, b, Canard, b. | | Deposit date: | 2014-06-23 | | Release date: | 2014-07-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | CRYSTAL STRUCTURE OF CHIKUNGUNYA VIRUS NSP3 MACRO DOMAIN IN COMPLEX WITH A 2'-5' OLIGOADENYLATE TRIMER
TO BE PUBLISHED
|
|
2CKW
 
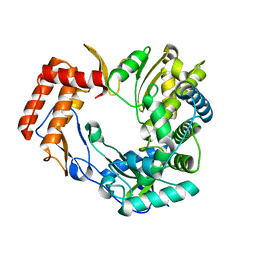 | | The 2.3 A resolution structure of the Sapporo virus RNA dependant RNA polymerase. | | Descriptor: | RNA-DIRECTED RNA POLYMERASE | | Authors: | Fullerton, S.W.B, Tucker, P.A, Rohayem, J, Coutard, B, Gebhardt, J, Gorbalenya, A, Canard, B. | | Deposit date: | 2006-04-24 | | Release date: | 2006-11-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and Functional Characterization of Sapovirus RNA-Dependent RNA Polymerase.
J.Virol., 81, 2007
|
|
2H85
 
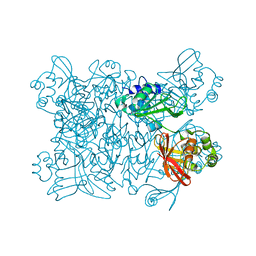 | | Crystal Structure of Nsp 15 from SARS | | Descriptor: | Putative orf1ab polyprotein | | Authors: | Ricagno, S, Egloff, M.P, Ulferts, R, Coutard, B, Nurizzo, D, Campanacci, V, Cambillau, C, Ziebuhr, J, Canard, B. | | Deposit date: | 2006-06-06 | | Release date: | 2006-08-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure and mechanistic determinants of SARS coronavirus nonstructural protein 15 define an endoribonuclease family.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
5LRP
 
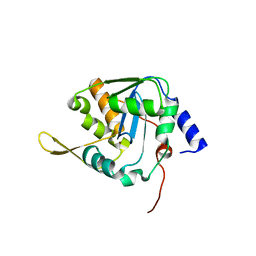 | | Mopeia Virus Exonuclease domain complexed with Magnesium | | Descriptor: | MAGNESIUM ION, Nucleoprotein, ZINC ION | | Authors: | Yekwa, E.L, Khourieh, J, Canard, B, Ferron, F. | | Deposit date: | 2016-08-19 | | Release date: | 2017-07-05 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.941 Å) | | Cite: | Activity inhibition and crystal polymorphism induced by active-site metal swapping.
Acta Crystallogr D Struct Biol, 73, 2017
|
|
5LTS
 
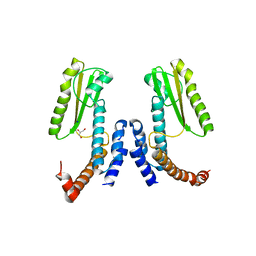 | | Crystal structure of Lymphocytic choriomeningitis mammarenavirus endonuclease Mutant D118A | | Descriptor: | CHLORIDE ION, GLYCEROL, RNA-directed RNA polymerase L, ... | | Authors: | Saez-Ayala, M, Yekwa, E.L, Canard, B, Alvarez, K, Ferron, F. | | Deposit date: | 2016-09-07 | | Release date: | 2017-09-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Crystal structures ofLymphocytic choriomeningitis virusendonuclease domain complexed with diketo-acid ligands.
IUCrJ, 5, 2018
|
|
5LS4
 
 | | Mopeia virus exonuclease domain complexed with Calcium | | Descriptor: | CALCIUM ION, DI(HYDROXYETHYL)ETHER, Nucleoprotein, ... | | Authors: | Yekwa, E.L, Canard, B, Ferron, F. | | Deposit date: | 2016-08-22 | | Release date: | 2017-07-05 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.469 Å) | | Cite: | Activity inhibition and crystal polymorphism induced by active-site metal swapping.
Acta Crystallogr D Struct Biol, 73, 2017
|
|
5LTN
 
 | | Crystal structure of Lymphocytic choriomeningitis mammarenavirus endonuclease complexed with DPBA | | Descriptor: | 2-4-DIOXO-4-PHENYLBUTANOIC ACID, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Saez-Ayala, M, Yekwa, E.L, Canard, B, Alvarez, K, Ferron, F. | | Deposit date: | 2016-09-07 | | Release date: | 2017-09-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Crystal structures of Lymphocytic choriomeningitis virusendonuclease domain complexed with diketo-acid ligands.
Iucrj, 5, 2018
|
|
5LTF
 
 | | Crystal structure of Lymphocytic choriomeningitis mammarenavirus endonuclease complexed with catalytic ions | | Descriptor: | MAGNESIUM ION, RNA-directed RNA polymerase L | | Authors: | Saez-Ayala, M, Yekwa, E.L, Canard, B, Ferron, F. | | Deposit date: | 2016-09-06 | | Release date: | 2017-09-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Crystal structures ofLymphocytic choriomeningitis virusendonuclease domain complexed with diketo-acid ligands.
IUCrJ, 5, 2018
|
|
3GPG
 
 | |
3GPO
 
 | | Crystal structure of macro domain of Chikungunya virus in complex with ADP-ribose | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, Non-structural protein 3 | | Authors: | Malet, H, Jamal, S, Coutard, B, Canard, B. | | Deposit date: | 2009-03-23 | | Release date: | 2009-07-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The crystal structures of Chikungunya and Venezuelan equine encephalitis virus nsP3 macro domains define a conserved adenosine binding pocket
J.Virol., 83, 2009
|
|
3GQE
 
 | | Crystal structure of macro domain of Venezuelan Equine Encephalitis virus | | Descriptor: | BICINE, Non-structural protein 3 | | Authors: | Jamal, S, Malet, H, Coutard, B, Canard, B. | | Deposit date: | 2009-03-24 | | Release date: | 2009-07-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The crystal structures of Chikungunya and Venezuelan equine encephalitis virus nsP3 macro domains define a conserved adenosine binding pocket
J.Virol., 83, 2009
|
|
3GQO
 
 | | Crystal structure of macro domain of Venezuelan Equine Encephalitis virus in complex with ADP-ribose | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, Non-structural protein 3 | | Authors: | Malet, H, Jamal, S, Coutard, B, Ferron, F, Canard, B. | | Deposit date: | 2009-03-24 | | Release date: | 2009-07-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The crystal structures of Chikungunya and Venezuelan equine encephalitis virus nsP3 macro domains define a conserved adenosine binding pocket
J.Virol., 83, 2009
|
|
