5HRQ
 
 | | Insulin with proline analog HzP at position B28 in the R6 state | | Descriptor: | CHLORIDE ION, Insulin A-Chain, Insulin B-Chain, ... | | Authors: | Lieblich, S.A, Fang, K.Y, Cahn, J.K.B, Tirrell, D.A. | | Deposit date: | 2016-01-24 | | Release date: | 2017-01-25 | | Last modified: | 2025-04-02 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | 4S-Hydroxylation of Insulin at ProB28 Accelerates Hexamer Dissociation and Delays Fibrillation.
J. Am. Chem. Soc., 139, 2017
|
|
5HPR
 
 | | Insulin with proline analog HyP at position B28 in the T2 state | | Descriptor: | GLYCEROL, Insulin A-Chain, Insulin B-Chain, ... | | Authors: | Lieblich, S.A, Fang, K.Y, Cahn, J.K.B, Tirrell, D.A. | | Deposit date: | 2016-01-21 | | Release date: | 2017-01-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | 4S-Hydroxylation of Insulin at ProB28 Accelerates Hexamer Dissociation and Delays Fibrillation.
J. Am. Chem. Soc., 139, 2017
|
|
5HQI
 
 | | Insulin with proline analog HzP at position B28 in the T2 state | | Descriptor: | Insulin A-Chain, Insulin B-Chain | | Authors: | Lieblich, S.A, Fang, K.Y, Cahn, J.K.B, Tirrell, D.A. | | Deposit date: | 2016-01-21 | | Release date: | 2017-01-25 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (0.97 Å) | | Cite: | 4S-Hydroxylation of Insulin at ProB28 Accelerates Hexamer Dissociation and Delays Fibrillation.
J. Am. Chem. Soc., 139, 2017
|
|
5HPU
 
 | | Insulin with proline analog HyP at position B28 in the R6 state | | Descriptor: | CHLORIDE ION, Insulin, chain A, ... | | Authors: | Lieblich, S.A, Fang, K.Y, Cahn, J.K.B, Tirrell, D.A. | | Deposit date: | 2016-01-21 | | Release date: | 2017-01-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | 4S-Hydroxylation of Insulin at ProB28 Accelerates Hexamer Dissociation and Delays Fibrillation.
J. Am. Chem. Soc., 139, 2017
|
|
6SBG
 
 | | Structure of type II terpene cyclase MstE_R337A from Scytonema in complex with geranylgeranyl dihydroxybenzoate (substrate) | | Descriptor: | GLYCEROL, MstE, geranylgeranyl dihydroxybenzoate | | Authors: | Moosmann, P, Ecker, F, Leopold-Messer, S, Cahn, J.K.B, Groll, M, Piel, J. | | Deposit date: | 2019-07-19 | | Release date: | 2020-07-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A monodomain class II terpene cyclase assembles complex isoprenoid scaffolds.
Nat.Chem., 12, 2020
|
|
6SBB
 
 | | Structure of type II terpene cyclase MstE from Scytonema (apo) | | Descriptor: | 1-ETHOXY-2-(2-ETHOXYETHOXY)ETHANE, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Moosmann, P, Ecker, F, Leopold-Messer, S, Cahn, J.K.B, Groll, M, Piel, J. | | Deposit date: | 2019-07-19 | | Release date: | 2020-07-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | A monodomain class II terpene cyclase assembles complex isoprenoid scaffolds.
Nat.Chem., 12, 2020
|
|
6SBF
 
 | | Structure of type II terpene cyclase MstE_Y157F from Scytonema (apo) | | Descriptor: | BETA-MERCAPTOETHANOL, GLYCEROL, MstE | | Authors: | Moosmann, P, Ecker, F, Leopold-Messer, S, Cahn, J.K.B, Groll, M, Piel, J. | | Deposit date: | 2019-07-19 | | Release date: | 2020-07-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | A monodomain class II terpene cyclase assembles complex isoprenoid scaffolds.
Nat.Chem., 12, 2020
|
|
6SBD
 
 | | Structure of type II terpene cyclase MstE_D109A from Scytonema in complex with merosterolic acid A (product) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, (4~{a}~{R},4~{b}~{S},6~{a}~{R},11~{a}~{R},11~{b}~{S},13~{a}~{R})-1,1,4~{a},6~{a},11~{b}-pentamethyl-9,10-bis(oxidanyl)- 3,4,4~{b},5,6,11,11~{a},12,13,13~{a}-decahydro-2~{H}-indeno[2,1-a]phenanthrene-7-carboxylic acid, GLYCEROL, ... | | Authors: | Moosmann, P, Ecker, F, Leopold-Messer, S, Cahn, J.K.B, Groll, M, Piel, J. | | Deposit date: | 2019-07-19 | | Release date: | 2020-07-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | A monodomain class II terpene cyclase assembles complex isoprenoid scaffolds.
Nat.Chem., 12, 2020
|
|
6SBC
 
 | | Structure of type II terpene cyclase MstE from Scytonema in complex with farnesyl dihydroxybenzoate | | Descriptor: | CHLORIDE ION, MstE, SODIUM ION, ... | | Authors: | Moosmann, P, Ecker, F, Leopold-Messer, S, Cahn, J.K.B, Groll, M, Piel, J. | | Deposit date: | 2019-07-19 | | Release date: | 2020-07-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | A monodomain class II terpene cyclase assembles complex isoprenoid scaffolds.
Nat.Chem., 12, 2020
|
|
6SBE
 
 | | Structure of type II terpene cyclase MstE_D109N from Scytonema in complex with geranylgeranyl dihydroxybenzoate (substrate) | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, MstE, ... | | Authors: | Moosmann, P, Ecker, F, Leopold-Messer, S, Cahn, J.K.B, Groll, M, Piel, J. | | Deposit date: | 2019-07-19 | | Release date: | 2020-07-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | A monodomain class II terpene cyclase assembles complex isoprenoid scaffolds.
Nat.Chem., 12, 2020
|
|
5BR4
 
 | | E. coli lactaldehyde reductase (FucO) M185C mutant | | Descriptor: | CHLORIDE ION, GLYCEROL, Lactaldehyde reductase, ... | | Authors: | Cahn, J.K.B, Brinkmann-Chen, S, Arnold, F.H. | | Deposit date: | 2015-05-29 | | Release date: | 2015-12-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (0.91 Å) | | Cite: | Mutations in adenine-binding pockets enhance catalytic properties of NAD(P)H-dependent enzymes.
Protein Eng.Des.Sel., 29, 2016
|
|
4XDZ
 
 | | Holo structure of ketol-acid reductoisomerase from Ignisphaera aggregans | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, GLYCEROL, Ketol-acid reductoisomerase, ... | | Authors: | Cahn, J.K.B, Brinkmann-Chen, S, Arnold, F.H. | | Deposit date: | 2014-12-20 | | Release date: | 2015-04-22 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Cofactor specificity motifs and the induced fit mechanism in class I ketol-acid reductoisomerases.
Biochem.J., 468, 2015
|
|
4XEH
 
 | |
4XDY
 
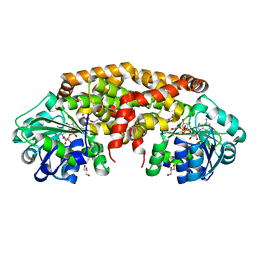 | | Structure of NADH-preferring ketol-acid reductoisomerase from an uncultured archean | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, GLYCEROL, Ketol-acid reductoisomerase, ... | | Authors: | Cahn, J.K.B, Brinkmann-Chen, S, Arnold, F.H. | | Deposit date: | 2014-12-20 | | Release date: | 2015-04-22 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.535 Å) | | Cite: | Cofactor specificity motifs and the induced fit mechanism in class I ketol-acid reductoisomerases.
Biochem.J., 468, 2015
|
|
5E4R
 
 | | Crystal structure of domain-duplicated synthetic class II ketol-acid reductoisomerase 2Ia_KARI-DD | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Ketol-acid reductoisomerase, ... | | Authors: | Cahn, J.K.B, Brinkmann-Chen, S, Buller, A.R, Arnold, F.H. | | Deposit date: | 2015-10-07 | | Release date: | 2015-12-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Artificial domain duplication replicates evolutionary history of ketol-acid reductoisomerases.
Protein Sci., 25, 2016
|
|
5D40
 
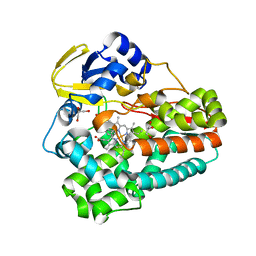 | | Crystal structure of the 5-selective H176Y mutant of Cytochrome TxtE | | Descriptor: | CHLORIDE ION, GLYCEROL, P450-like protein, ... | | Authors: | Cahn, J.K.B, Dodani, S.C, Arnold, F.H. | | Deposit date: | 2015-08-06 | | Release date: | 2016-06-22 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Discovery of a regioselectivity switch in nitrating P450s guided by molecular dynamics simulations and Markov models.
Nat.Chem., 8, 2016
|
|
5D3U
 
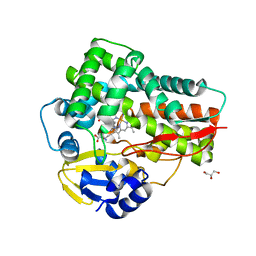 | | Crystal structure of the 5-selective H176F mutant of Cytochrome TxtE | | Descriptor: | CHLORIDE ION, GLYCEROL, P450-like protein, ... | | Authors: | Cahn, J.K.B, Dodani, S.C, Arnold, F.H. | | Deposit date: | 2015-08-06 | | Release date: | 2016-06-22 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Discovery of a regioselectivity switch in nitrating P450s guided by molecular dynamics simulations and Markov models.
Nat.Chem., 8, 2016
|
|
4TPO
 
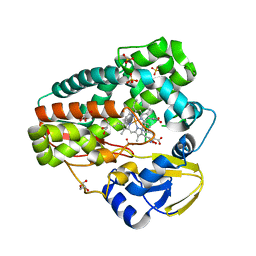 | | High-resolution structure of TxtE with bound tryptophan substrate | | Descriptor: | GLYCEROL, PROTOPORPHYRIN IX CONTAINING FE, Putative P450-like protein, ... | | Authors: | Cahn, J.K.B, Dodani, S.C, Brinkmann-Chen, S, Heinsich, T, McIntosh, J.A, Arnold, F.H. | | Deposit date: | 2014-06-08 | | Release date: | 2014-09-10 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.225 Å) | | Cite: | Structural, Functional, and Spectroscopic Characterization of the Substrate Scope of the Novel Nitrating Cytochrome P450 TxtE.
Chembiochem, 15, 2014
|
|
4TPN
 
 | | High-resolution structure of TxtE in the absence of substrate | | Descriptor: | GLYCEROL, LITHIUM ION, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Cahn, J.K.B, Dodani, S.C, Brinkmann-Chen, S, Heinsich, T, McIntosh, J.A, Arnold, F.H. | | Deposit date: | 2014-06-08 | | Release date: | 2014-09-10 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.18 Å) | | Cite: | Structural, Functional, and Spectroscopic Characterization of the Substrate Scope of the Novel Nitrating Cytochrome P450 TxtE.
Chembiochem, 15, 2014
|
|
4TSK
 
 | | Ketol-acid reductoisomerase from Alicyclobacillus acidocaldarius | | Descriptor: | Ketol-acid reductoisomerase, L(+)-TARTARIC ACID, MAGNESIUM ION, ... | | Authors: | Cahn, J.K.B, Brinkmann-Chen, S, Arnold, F.H. | | Deposit date: | 2014-06-18 | | Release date: | 2014-07-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Uncovering rare NADH-preferring ketol-acid reductoisomerases.
Metab. Eng., 26C, 2014
|
|
4KQX
 
 | | Mutant Slackia exigua KARI DDV in complex with NAD and an inhibitor | | Descriptor: | HISTIDINE, Ketol-acid reductoisomerase, MAGNESIUM ION, ... | | Authors: | Brinkmann-Chen, S, Flock, T, Cahn, J.K.B, Snow, C.D, Brustad, E.M, Mcintosh, J.A, Meinhold, P, Zhang, L, Arnold, F.H. | | Deposit date: | 2013-05-15 | | Release date: | 2013-06-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | General approach to reversing ketol-acid reductoisomerase cofactor dependence from NADPH to NADH.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4KQW
 
 | | The structure of the Slackia exigua KARI in complex with NADP | | Descriptor: | Ketol-acid reductoisomerase, L(+)-TARTARIC ACID, MAGNESIUM ION, ... | | Authors: | Brinkmann-Chen, S, Flock, T, Cahn, J.K.B, Snow, C.D, Brustad, E.M, Mcintosh, J.A, Meinhold, P, Zhang, L, Arnold, F.H. | | Deposit date: | 2013-05-15 | | Release date: | 2013-06-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | General approach to reversing ketol-acid reductoisomerase cofactor dependence from NADPH to NADH.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4XIY
 
 | | Crystal structure of ketol-acid reductoisomerase from Azotobacter | | Descriptor: | DI(HYDROXYETHYL)ETHER, FE (III) ION, Ketol-acid reductoisomerase, ... | | Authors: | Spatzal, T, Cahn, J.K.B, Wiig, J.A, Einsle, O, Hu, Y, Ribbe, M.W, Arnold, F.H. | | Deposit date: | 2015-01-07 | | Release date: | 2015-04-22 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Cofactor specificity motifs and the induced fit mechanism in class I ketol-acid reductoisomerases.
Biochem.J., 468, 2015
|
|
5UU8
 
 | |
5UUD
 
 | | Tetragonal thermolysin cryocooled to 100 K with 50% dmf as cryoprotectant | | Descriptor: | CALCIUM ION, CHLORIDE ION, DIMETHYLFORMAMIDE, ... | | Authors: | Juers, D.H. | | Deposit date: | 2017-02-16 | | Release date: | 2017-03-08 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.60000348 Å) | | Cite: | The impact of cryosolution thermal contraction on proteins and protein crystals: volumes, conformation and order.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
