7K65
 
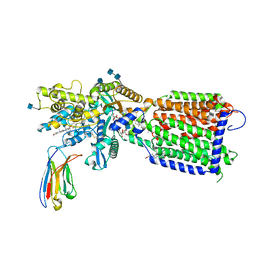 | | Hedgehog receptor Patched (PTCH1) in complex with conformation selective nanobody TI23 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-{[(4-O-alpha-D-glucopyranosyl-alpha-D-glucopyranosyl)oxy]methyl}-4-{[(3beta,9beta,14beta,17beta,25R)-spirost-5-en-3-yl]oxy}butyl 4-O-alpha-D-glucopyranosyl-alpha-D-glucopyranoside, ... | | Authors: | Zhang, Y, Bulkley, D.P, Liang, J, Manglik, A, Cheng, Y, Beachy, P.A. | | Deposit date: | 2020-09-18 | | Release date: | 2021-03-17 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Hedgehog pathway activation through nanobody-mediated conformational blockade of the Patched sterol conduit.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
4V7Y
 
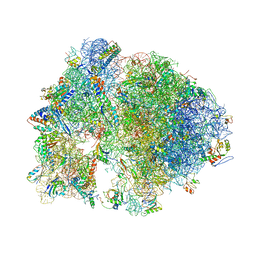 | | Structure of the Thermus thermophilus 70S ribosome complexed with azithromycin. | | Descriptor: | 16S rRNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Bulkley, D.P, Innis, C.A, Blaha, G, Steitz, T.A. | | Deposit date: | 2010-08-18 | | Release date: | 2014-07-09 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Revisiting the structures of several antibiotics bound to the bacterial ribosome.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
4V7W
 
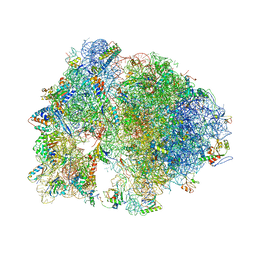 | | Structure of the Thermus thermophilus ribosome complexed with chloramphenicol. | | Descriptor: | 16S rRNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Bulkley, D.P, Innis, C.A, Blaha, G, Steitz, T.A. | | Deposit date: | 2010-08-16 | | Release date: | 2014-07-09 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Revisiting the structures of several antibiotics bound to the bacterial ribosome.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
4V9R
 
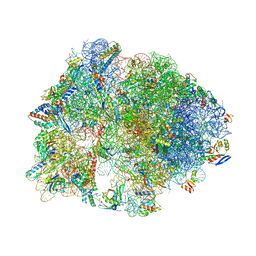 | | Crystal structure of antibiotic DITYROMYCIN bound to 70S ribosome | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S Ribosomal Protein S10, ... | | Authors: | Bulkley, D.P, Brandi, L, Polikanov, Y.S, Fabbretti, A, O'Connor, M, Gualerzi, C.O, Steitz, T.A. | | Deposit date: | 2013-12-05 | | Release date: | 2014-07-09 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The antibiotics dityromycin and GE82832 bind protein S12 and block EF-G-catalyzed translocation.
Cell Rep, 6, 2014
|
|
4V7Z
 
 | | Structure of the Thermus thermophilus 70S ribosome complexed with telithromycin. | | Descriptor: | 16S rRNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Bulkley, D.P, Innis, C.A, Blaha, G, Steitz, T.A. | | Deposit date: | 2010-08-18 | | Release date: | 2014-07-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Revisiting the structures of several antibiotics bound to the bacterial ribosome.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
4V7X
 
 | | Structure of the Thermus thermophilus ribosome complexed with erythromycin. | | Descriptor: | 16S rRNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Bulkley, D.P, Innis, C.A, Blaha, G, Steitz, T.A. | | Deposit date: | 2010-08-17 | | Release date: | 2014-07-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Revisiting the structures of several antibiotics bound to the bacterial ribosome.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
4V9S
 
 | | Crystal structure of antibiotic GE82832 bound to 70S ribosome | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S Ribosomal Protein S10, ... | | Authors: | Bulkley, D.P, Brandi, L, Polikanov, Y.S, Fabbretti, A, O'Connor, M, Gualerzi, C.O, Steitz, T.A. | | Deposit date: | 2013-12-05 | | Release date: | 2014-07-09 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | The antibiotics dityromycin and GE82832 bind protein S12 and block EF-G-catalyzed translocation.
Cell Rep, 6, 2014
|
|
6MG8
 
 | | Structural basis for cholesterol transport-like activity of the Hedgehog receptor Patched | | Descriptor: | CHOLESTEROL, Protein patched homolog 1 | | Authors: | Zhang, Y, Bulkley, D, Xin, Y, Roberts, K.J, Asarnow, D.E, Sharma, A, Myers, B.R, Cho, W, Cheng, Y, Beachy, P.A. | | Deposit date: | 2018-09-13 | | Release date: | 2018-11-28 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural Basis for Cholesterol Transport-like Activity of the Hedgehog Receptor Patched.
Cell, 175, 2018
|
|
4V95
 
 | | Crystal structure of YAEJ bound to the 70S ribosome | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S Ribosomal Protein S10, ... | | Authors: | Gagnon, M.G, Seetharaman, S.V, Bulkley, D.P, Steitz, T.A. | | Deposit date: | 2012-01-27 | | Release date: | 2014-07-09 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural basis for the rescue of stalled ribosomes: structure of YaeJ bound to the ribosome.
Science, 335, 2012
|
|
6UFI
 
 | | W96Y Oxalate Decarboxylase (Bacillus subtilis) | | Descriptor: | CHLORIDE ION, Cupin domain-containing protein, GLYCEROL, ... | | Authors: | Pastore, A.J, Burg, M.J, Twahir, U.T, Bruner, S.D, Angerhofer, A. | | Deposit date: | 2019-09-24 | | Release date: | 2020-09-30 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Oxalate decarboxylase uses electron hole hopping for catalysis.
J.Biol.Chem., 297, 2021
|
|
6TZP
 
 | | W96F Oxalate Decarboxylase (B. subtilis) | | Descriptor: | MANGANESE (II) ION, Oxalate decarboxylase | | Authors: | Pastore, A.J, Burg, M.J, Twahir, U.T, Bruner, S.D, Angerhofer, A. | | Deposit date: | 2019-08-12 | | Release date: | 2021-02-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Oxalate decarboxylase uses electron hole hopping for catalysis.
J.Biol.Chem., 297, 2021
|
|
7KOD
 
 | | Cryo-EM structure of heavy chain mouse apoferritin | | Descriptor: | Ferritin heavy chain | | Authors: | Sun, M, Azumaya, C, Tse, E, Frost, A, Southworth, D, Verba, K.A, Cheng, Y, Agard, D.A. | | Deposit date: | 2020-11-08 | | Release date: | 2020-12-16 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (1.655 Å) | | Cite: | Practical considerations for using K3 cameras in CDS mode for high-resolution and high-throughput single particle cryo-EM.
J.Struct.Biol., 213, 2021
|
|
