2L99
 
 | | Solution structure of LAK160-P10 | | Descriptor: | LAK160-P10 | | Authors: | Vermeer, L.S, Bui, T.T, Lan, Y, Jumagulova, E, Kozlowska, J, McIntyre, C, Drake, A.F, Mason, J.A. | | Deposit date: | 2011-02-03 | | Release date: | 2012-02-01 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | The role of proline induced conformational flexibility in determining the antibacterial potency of linear cationic alpha-helical peptides
To be Published
|
|
2L9A
 
 | | Solution structure of LAK160-P12 | | Descriptor: | LAK160-P12 | | Authors: | Vermeer, L.S, Bui, T.T, Lan, Y, Jumagulova, E, Kozlowska, J, McIntyre, C, Drake, A.F, Mason, J.A. | | Deposit date: | 2011-02-03 | | Release date: | 2012-02-01 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | The role of proline induced conformational flexibility in determining the antibacterial potency of linear cationic alpha-helical peptides
To be Published
|
|
2L96
 
 | | Solution structure of LAK160-P7 | | Descriptor: | LAK160-P7 | | Authors: | Vermeer, L.S, Bui, T.T, Lan, Y, Jumagulova, E, Kozlowska, J, McIntyre, C, Drake, A.F, Mason, J.A. | | Deposit date: | 2011-02-01 | | Release date: | 2012-02-01 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | The role of proline induced conformational flexibility in determining the antibacterial potency of linear cationic alpha-helical peptides
To be Published
|
|
2MTG
 
 | | Solution structure of the RRM1 of human LARP6 | | Descriptor: | La-related protein 6 | | Authors: | Martino, L, Atkinson, A.R, Kelly, G, Conte, M.R. | | Deposit date: | 2014-08-18 | | Release date: | 2014-12-24 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Synergic interplay of the La motif, RRM1 and the interdomain linker of LARP6 in the recognition of collagen mRNA expands the RNA binding repertoire of the La module.
Nucleic Acids Res., 43, 2015
|
|
2MTF
 
 | | Solution structure of the La motif of human LARP6 | | Descriptor: | La-related protein 6 | | Authors: | Martino, L, Salisbury, N.JH, Atkinson, A.R, Kelly, G, Conte, M.R. | | Deposit date: | 2014-08-18 | | Release date: | 2014-12-24 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Synergic interplay of the La motif, RRM1 and the interdomain linker of LARP6 in the recognition of collagen mRNA expands the RNA binding repertoire of the La module.
Nucleic Acids Res., 43, 2015
|
|
6RSG
 
 | | NMR structure of pleurocidin VA in SDS micelles | | Descriptor: | Pleurocidin | | Authors: | Manzo, G, Mason, A.J. | | Deposit date: | 2019-05-21 | | Release date: | 2020-12-09 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | A pleurocidin analogue with greater conformational flexibility, enhanced antimicrobial potency and in vivo therapeutic efficacy.
Commun Biol, 3, 2020
|
|
6RSF
 
 | | NMR structure of pleurocidin KR in SDS micelles | | Descriptor: | Pleurocidin | | Authors: | Manzo, G, Mason, A.J. | | Deposit date: | 2019-05-21 | | Release date: | 2020-12-09 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | A pleurocidin analogue with greater conformational flexibility, enhanced antimicrobial potency and in vivo therapeutic efficacy.
Commun Biol, 3, 2020
|
|
6GIK
 
 | | NMR structure of temporin B L1FK in SDS micelles | | Descriptor: | temporinB_L1FK | | Authors: | Manzo, G, Mason, J.A. | | Deposit date: | 2018-05-12 | | Release date: | 2018-06-13 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Minor sequence modifications in temporin B cause drastic changes in antibacterial potency and selectivity by fundamentally altering membrane activity.
Sci Rep, 9, 2019
|
|
6GIL
 
 | | NMR structure of temporin B in SDS micelles | | Descriptor: | Temporin-B | | Authors: | Manzo, G, Mason, J.A. | | Deposit date: | 2018-05-12 | | Release date: | 2018-06-13 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Minor sequence modifications in temporin B cause drastic changes in antibacterial potency and selectivity by fundamentally altering membrane activity.
Sci Rep, 9, 2019
|
|
6GS5
 
 | | NMR structure of temporin L in SDS micelles | | Descriptor: | Temporin-L | | Authors: | Manzo, G, Mason, J.A. | | Deposit date: | 2018-06-13 | | Release date: | 2018-07-18 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Temporin L and aurein 2.5 have identical conformations but subtly distinct membrane and antibacterial activities.
Sci Rep, 9, 2019
|
|
6GS9
 
 | | NMR structure of aurein 2.5 in SDS micelles | | Descriptor: | Aurein 2.5 | | Authors: | Manzo, G, Mason, J.A. | | Deposit date: | 2018-06-13 | | Release date: | 2018-07-18 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Temporin L and aurein 2.5 have identical conformations but subtly distinct membrane and antibacterial activities.
Sci Rep, 9, 2019
|
|
6GIJ
 
 | | NMR structure of temporin B KKG6A in SDS micelles | | Descriptor: | temporinB_KKG6A | | Authors: | Manzo, G, Mason, J.A. | | Deposit date: | 2018-05-12 | | Release date: | 2018-06-13 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Minor sequence modifications in temporin B cause drastic changes in antibacterial potency and selectivity by fundamentally altering membrane activity.
Sci Rep, 9, 2019
|
|
4E68
 
 | | Unphosphorylated STAT3B core protein binding to dsDNA | | Descriptor: | DNA (5'-D(*TP*GP*CP*AP*TP*TP*TP*CP*CP*CP*GP*TP*AP*AP*AP*TP*CP*T)-3'), Signal transducer and activator of transcription 3 | | Authors: | Collie, G.W, Parkinson, G.N, Shah, R. | | Deposit date: | 2012-03-15 | | Release date: | 2013-03-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.585 Å) | | Cite: | Observation of unphosphorylated STAT3 core protein binding to target dsDNA by PEMSA and X-ray crystallography.
Febs Lett., 587, 2013
|
|
6O8S
 
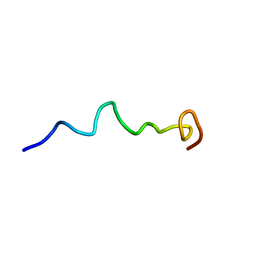 | | Syn-safencin 56 | | Descriptor: | Circular bacteriocin, circularin A/uberolysin family | | Authors: | Fields, F.R, Lee, S.W. | | Deposit date: | 2019-03-11 | | Release date: | 2020-05-13 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Synthetic Antimicrobial Peptide Tuning Permits Membrane Disruption and Interpeptide Synergy.
Acs Pharmacol Transl Sci, 3, 2020
|
|
6O8T
 
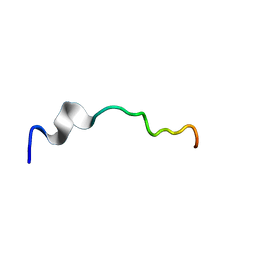 | | Syn-safencin 96 | | Descriptor: | Circular bacteriocin, circularin A/uberolysin family | | Authors: | Fields, F.R, Lee, S.W. | | Deposit date: | 2019-03-11 | | Release date: | 2020-04-15 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Synthetic Antimicrobial Peptide Tuning Permits Membrane Disruption and Interpeptide Synergy.
Acs Pharmacol Transl Sci, 3, 2020
|
|
6O8R
 
 | | Syn-safencin 24 | | Descriptor: | Circular bacteriocin, circularin A/uberolysin family | | Authors: | Fields, F.R, Lee, S.W. | | Deposit date: | 2019-03-11 | | Release date: | 2020-05-13 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Synthetic Antimicrobial Peptide Tuning Permits Membrane Disruption and Interpeptide Synergy.
Acs Pharmacol Transl Sci, 3, 2020
|
|
6O8J
 
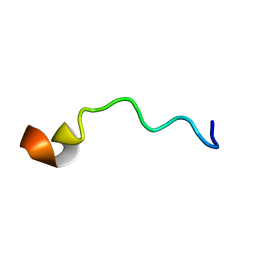 | | Syn-safencin | | Descriptor: | Circular bacteriocin, circularin A/uberolysin family | | Authors: | Fields, F.R, Lee, S.W. | | Deposit date: | 2019-03-11 | | Release date: | 2020-04-15 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Synthetic Antimicrobial Peptide Tuning Permits Membrane Disruption and Interpeptide Synergy.
Acs Pharmacol Transl Sci, 3, 2020
|
|
6O8P
 
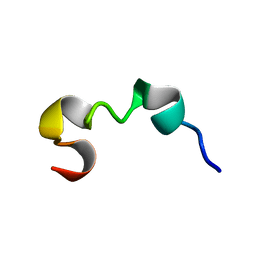 | | Syn-safencin 8 | | Descriptor: | Circular bacteriocin, circularin A/uberolysin family | | Authors: | Fields, F.R, Lee, S.W. | | Deposit date: | 2019-03-11 | | Release date: | 2020-05-13 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Synthetic Antimicrobial Peptide Tuning Permits Membrane Disruption and Interpeptide Synergy.
Acs Pharmacol Transl Sci, 3, 2020
|
|
6I5G
 
 | | X-ray structure of human soluble Epoxide Hydrolase C-terminal Domain (hsEH CTD)in complex with 15d-PGJ2 | | Descriptor: | (5E,14E)-11-oxoprosta-5,9,12,14-tetraen-1-oic acid, 1,2-ETHANEDIOL, Bifunctional epoxide hydrolase 2 | | Authors: | Abis, G, Kopec, J, Yue, W.W, Conte, M.R. | | Deposit date: | 2018-11-13 | | Release date: | 2019-05-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | 15-deoxy-Delta12,14-Prostaglandin J2inhibits human soluble epoxide hydrolase by a dual orthosteric and allosteric mechanism.
Commun Biol, 2, 2019
|
|
6I5E
 
 | | X-ray structure of apo human soluble Epoxide Hydrolase C-terminal Domain (hsEH CTD) | | Descriptor: | Bifunctional epoxide hydrolase 2 | | Authors: | Abis, G, Kopec, J, Yue, W.W, Conte, M.R. | | Deposit date: | 2018-11-13 | | Release date: | 2019-05-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | 15-deoxy-Delta12,14-Prostaglandin J2inhibits human soluble epoxide hydrolase by a dual orthosteric and allosteric mechanism.
Commun Biol, 2, 2019
|
|
6I9B
 
 | | NMR structure of the La module from human LARP4A | | Descriptor: | La-related protein 4 | | Authors: | Conte, M.R, Martino, L, Atkinson, R.A, Kelly, G, Cruz-Gallardo, I, De Tito, S, Trotta, R. | | Deposit date: | 2018-11-22 | | Release date: | 2019-03-13 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | LARP4A recognizes polyA RNA via a novel binding mechanism mediated by disordered regions and involving the PAM2w motif, revealing interplay between PABP, LARP4A and mRNA.
Nucleic Acids Res., 47, 2019
|
|
