1XC8
 
 | | CRYSTAL STRUCTURE COMPLEX BETWEEN THE WILD-TYPE LACTOCOCCUS LACTIS FPG (MUTM) AND A FAPY-DG CONTAINING DNA | | Descriptor: | 5'-D(*CP*TP*CP*TP*TP*TP*(FOX)P*TP*TP*TP*CP*TP*CP*G)-3', 5'-D(*GP*CP*GP*AP*GP*AP*AP*AP*CP*AP*AP*AP*GP*A)-3', Formamidopyrimidine-DNA glycosylase, ... | | Authors: | Coste, F, Ober, M, Carell, T, Boiteux, S, Zelwer, C, Castaing, B. | | Deposit date: | 2004-09-01 | | Release date: | 2005-09-06 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural basis for the recognition of the FapydG lesion (2,6-diamino-4-hydroxy-5-formamidopyrimidine) by formamidopyrimidine-DNA glycosylase.
J.Biol.Chem., 279, 2004
|
|
1KFV
 
 | | Crystal Structure of Lactococcus lactis Formamido-pyrimidine DNA Glycosylase (alias Fpg or MutM) Non Covalently Bound to an AP Site Containing DNA. | | Descriptor: | 5'-D(*CP*TP*CP*TP*TP*TP*(PDI)P*TP*TP*TP*CP*TP*C)-3', 5'-D(*GP*AP*GP*AP*AP*AP*CP*AP*AP*AP*GP*AP*G)-3', Formamido-pyrimidine DNA glycosylase, ... | | Authors: | Serre, L, Pereira de Jesus, K, Boiteux, S, Zelwer, C, Castaing, B. | | Deposit date: | 2001-11-23 | | Release date: | 2002-06-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Crystal structure of the Lactococcus lactis formamidopyrimidine-DNA glycosylase bound to an abasic site analogue-containing DNA.
EMBO J., 21, 2002
|
|
1TDZ
 
 | | Crystal Structure Complex Between the Lactococcus Lactis FPG (Mutm) and a FAPY-dG Containing DNA | | Descriptor: | 5'-D(*CP*TP*CP*TP*TP*TP*(FOX)P*TP*TP*TP*CP*TP*CP*G)-3', 5'-D(*GP*CP*GP*AP*GP*AP*AP*AP*CP*AP*AP*AP*GP*A)-3', GLYCEROL, ... | | Authors: | Coste, F, Ober, M, Carell, T, Boiteux, S, Zelwer, C, Castaing, B. | | Deposit date: | 2004-05-24 | | Release date: | 2004-09-07 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for the recognition of the FapydG lesion (2,6-diamino-4-hydroxy-5-formamidopyrimidine) by formamidopyrimidine-DNA glycosylase
J.Biol.Chem., 279, 2004
|
|
4FMN
 
 | | Structure of the C-terminal domain of the Saccharomyces cerevisiae MUTL alpha (MLH1/PMS1) heterodimer bound to a fragment of NTG2 | | Descriptor: | 1,2-ETHANEDIOL, DNA mismatch repair protein MLH1, DNA mismatch repair protein PMS1, ... | | Authors: | Gueneau, E, Legrand, P, Charbonnier, J.B. | | Deposit date: | 2012-06-18 | | Release date: | 2013-02-20 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Structure of the MutL alpha C-terminal domain reveals how Mlh1 contributes to Pms1 endonuclease site.
Nat.Struct.Mol.Biol., 20, 2013
|
|
4FMO
 
 | | Structure of the C-terminal domain of the Saccharomyces cerevisiae MUTL alpha (MLH1/PMS1) heterodimer bound to a fragment of exo1 | | Descriptor: | DNA mismatch repair protein MLH1, DNA mismatch repair protein PMS1, DNA repair peptide, ... | | Authors: | Gueneau, E, Legrand, P, Charbonnier, J.B. | | Deposit date: | 2012-06-18 | | Release date: | 2013-02-20 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.04 Å) | | Cite: | Structure of the MutL alpha C-terminal domain reveals how Mlh1 contributes to Pms1 endonuclease site.
Nat.Struct.Mol.Biol., 20, 2013
|
|
4E4W
 
 | | Structure of the C-terminal domain of the Saccharomyces cerevisiae MUTL alpha (MLH1/PMS1) heterodimer | | Descriptor: | 1,2-ETHANEDIOL, DNA mismatch repair protein MLH1, DNA mismatch repair protein PMS1, ... | | Authors: | Gueneau, E, Legrand, P, Charbonnier, J.B. | | Deposit date: | 2012-03-13 | | Release date: | 2013-02-20 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of the MutLalpha C-terminal domain reveals how Mlh1 contributes to Pms1 endonuclease site.
Nat.Struct.Mol.Biol., 20, 2013
|
|
1NNJ
 
 | | Crystal structure Complex between the Lactococcus lactis Fpg and an abasic site containing DNA | | Descriptor: | 5'-D(*CP*TP*CP*TP*TP*TP*(PDI)P*TP*TP*TP*CP*TP*CP*G)-3', 5'-D(*GP*CP*GP*AP*GP*AP*AP*AP*CP*AP*AP*AP*GP*A)-3', Formamidopyrimidine-DNA glycosylase, ... | | Authors: | Serre, L, Pereira de Jesus, K, Boiteux, S, Zelwer, C, Castaing, B. | | Deposit date: | 2003-01-14 | | Release date: | 2003-02-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural insights into abasic site for Fpg specific binding and catalysis: comparative high-resolution crystallographic studies of Fpg bound to various models of abasic site analogues-containing DNA.
Nucleic Acids Res., 33, 2005
|
|
1PU8
 
 | | Crystal structure of H.pylori 3-methyladenine DNA glycosylase (MagIII) bound to 1,N6-ethenoadenine | | Descriptor: | 3-METHYLADENINE DNA GLYCOSYLASE, 3H-IMIDAZO[2,1-I]PURINE, BETA-MERCAPTOETHANOL | | Authors: | Eichman, B.F, O'Rourke, E.J, Radicella, J.P, Ellenberger, T. | | Deposit date: | 2003-06-24 | | Release date: | 2003-10-07 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Crystal structures of 3-methyladenine DNA glycosylase MagIII and the recognition of alkylated bases
Embo J., 22, 2003
|
|
1PM5
 
 | | Crystal structure of wild type Lactococcus lactis Fpg complexed to a tetrahydrofuran containing DNA | | Descriptor: | DNA (5'-D(*CP*TP*CP*TP*TP*TP*(3DR)P*TP*TP*TP*CP*TP*CP*G)-3'), DNA (5'-D(*GP*CP*GP*AP*GP*AP*AP*AP*CP*AP*AP*AP*GP*A)-3'), Formamidopyrimidine-DNA glycosylase, ... | | Authors: | Pereira de Jesus-Tran, K, Serre, L, Zelwer, C, Castaing, B. | | Deposit date: | 2003-06-10 | | Release date: | 2004-07-27 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural insights into abasic site for Fpg specific binding and catalysis: comparative high-resolution crystallographic studies of Fpg bound to various models of abasic site analogues-containing DNA.
Nucleic Acids Res., 33, 2005
|
|
1PU6
 
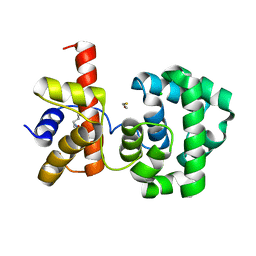 | | Crystal structure of H.pylori 3-methyladenine DNA glycosylase (MagIII) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 3-METHYLADENINE DNA GLYCOSYLASE, BETA-MERCAPTOETHANOL, ... | | Authors: | Eichman, B.F, O'Rourke, E.J, Radicella, J.P, Ellenberger, T. | | Deposit date: | 2003-06-24 | | Release date: | 2003-10-07 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Crystal structures of 3-methyladenine DNA glycosylase MagIII and the recognition of alkylated bases
Embo J., 22, 2003
|
|
1PU7
 
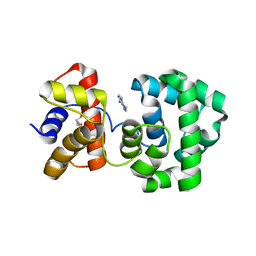 | | Crystal structure of H.pylori 3-methyladenine DNA glycosylase (MagIII) bound to 3,9-dimethyladenine | | Descriptor: | 3-METHYLADENINE DNA GLYCOSYLASE, 6-AMINO-3,9-DIMETHYL-9H-PURIN-3-IUM, BETA-MERCAPTOETHANOL | | Authors: | Eichman, B.F, O'Rourke, E.J, Radicella, J.P, Ellenberger, T. | | Deposit date: | 2003-06-24 | | Release date: | 2003-10-07 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Crystal structures of 3-methyladenine DNA glycosylase MagIII and the recognition of alkylated bases
Embo J., 22, 2003
|
|
1PJJ
 
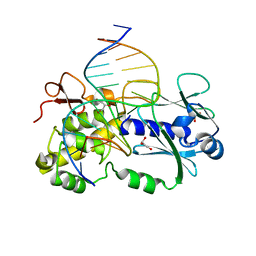 | | Complex between the Lactococcus lactis Fpg and an abasic site containing DNA. | | Descriptor: | DNA (5'-D(*CP*TP*CP*TP*TP*TP*(3DR)P*TP*TP*TP*CP*TP*CP*G)-3'), DNA (5'-D(*GP*CP*GP*AP*GP*AP*AP*AP*CP*AP*AP*AP*GP*A)-3'), Formamidopyrimidine-DNA glycosylase, ... | | Authors: | Serre, L, Pereira de Jesus, K, Boiteux, S, Zelwer, C, Castaing, B. | | Deposit date: | 2003-06-03 | | Release date: | 2004-06-08 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural insights into abasic site for Fpg specific binding and catalysis: comparative high-resolution crystallographic studies of Fpg bound to various models of abasic site analogues-containing DNA.
Nucleic Acids Res., 33, 2005
|
|
