6W3D
 
 | | Rd1NTF2_05 with long sheet | | Descriptor: | Rd1NTF2_05 | | Authors: | Bick, M.J, Basanta, B, Sankaran, B, Baker, D. | | Deposit date: | 2020-03-09 | | Release date: | 2020-04-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | An enumerative algorithm for de novo design of proteins with diverse pocket structures.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6W3G
 
 | | Rd1NTF2_04 with long sheet | | Descriptor: | Rd1NTF2_04 | | Authors: | Bick, M.J, Basanta, B, Sankaran, B, Baker, D. | | Deposit date: | 2020-03-09 | | Release date: | 2020-04-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | An enumerative algorithm for de novo design of proteins with diverse pocket structures.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6W2W
 
 | | Junction 24, DHR14-DHR18 | | Descriptor: | Junction 24 DHR14-DHR18 | | Authors: | Bick, M.J, Brunette, T.J, Baker, D. | | Deposit date: | 2020-03-08 | | Release date: | 2020-04-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Modular repeat protein sculpting using rigid helical junctions.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6W2R
 
 | | Junction 19, DHR54-DHR79 | | Descriptor: | Junction 19 DHR54-DHR79 | | Authors: | Bick, M.J, Brunette, T.J, Baker, D. | | Deposit date: | 2020-03-08 | | Release date: | 2020-04-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.344 Å) | | Cite: | Modular repeat protein sculpting using rigid helical junctions.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6W3F
 
 | |
6WVS
 
 | | Hyperstable de novo TIM barrel variant DeNovoTIM15 | | Descriptor: | DeNovoTIM15 hyperstable de novo TIM barrel | | Authors: | Bick, M.J, Haydon, I.C, Caldwell, S.J, Zeymer, C, Huang, P, Fernandez-Velasco, D.A, Baker, D. | | Deposit date: | 2020-05-06 | | Release date: | 2020-11-25 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.202 Å) | | Cite: | Tight and specific lanthanide binding in a de novo TIM barrel with a large internal cavity designed by symmetric domain fusion.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
4IJB
 
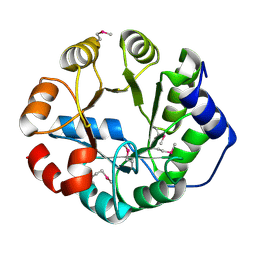 | | Crystal Structure of Engineered Protein, Northeast Structural Genomics Consortium Target OR288 | | Descriptor: | Engineered Protein OR288 | | Authors: | Vorobiev, S, Su, M, Bick, M.J, Seetharaman, J, Khare, S, Maglaqui, M, Xiao, R, Lee, D, Day, A, Everett, J.K, Acton, T.B, Baker, D, Montelione, G.T, Hunt, J, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2012-12-21 | | Release date: | 2013-01-16 | | Last modified: | 2013-02-27 | | Method: | X-RAY DIFFRACTION (1.739 Å) | | Cite: | Crystal Structure of Engineered Protein OR288.
To be Published
|
|
6MRR
 
 | | De novo designed protein Foldit1 | | Descriptor: | Foldit1 | | Authors: | Koepnick, B, Bick, M.J, Estep, R.D, Kleinfelter, S, Wei, L, Baker, D. | | Deposit date: | 2018-10-15 | | Release date: | 2019-06-12 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.18 Å) | | Cite: | De novo protein design by citizen scientists.
Nature, 570, 2019
|
|
6MSQ
 
 | | Crystal structure of pRO-2.3 | | Descriptor: | pRO-2.3 | | Authors: | Boyken, S.E, Sankaran, B, Bick, M.J, Baker, D. | | Deposit date: | 2018-10-17 | | Release date: | 2019-05-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | De novo design of tunable, pH-driven conformational changes.
Science, 364, 2019
|
|
3NRX
 
 | | Insights into anti-parallel microtubule crosslinking by PRC1, a conserved non-motor microtubule binding protein | | Descriptor: | Protein regulator of cytokinesis 1 | | Authors: | Kapoor, T.M, Subramanian, R, Wilson-Kubalek, E.M, Arthur, C.P, Bick, M.J, Campbell, E.A, Darst, S.A, Milligan, R.A. | | Deposit date: | 2010-06-30 | | Release date: | 2010-08-18 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Insights into Antiparallel Microtubule Crosslinking by PRC1, a Conserved Nonmotor Microtubule Binding Protein.
Cell(Cambridge,Mass.), 142, 2010
|
|
3NRY
 
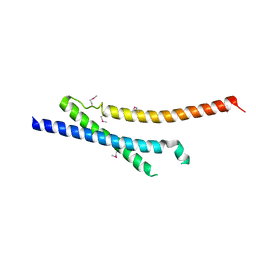 | | Insights into anti-parallel microtubule crosslinking by PRC1, a conserved microtubule binding protein | | Descriptor: | Protein regulator of cytokinesis 1 | | Authors: | Kapoor, T.M, Subramanian, R, Wilson-Kubalek, E.M, Arthur, C.P, Bick, M.J, Campbell, E.A, Darst, S.A, Milligan, R.A. | | Deposit date: | 2010-07-01 | | Release date: | 2010-08-25 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Insights into Antiparallel Microtubule Crosslinking by PRC1, a Conserved Nonmotor Microtubule Binding Protein.
Cell(Cambridge,Mass.), 142, 2010
|
|
6D0T
 
 | | De novo design of a fluorescence-activating beta barrel - BB1 | | Descriptor: | BB1 | | Authors: | Dou, J, Vorobieva, A.A, Sheffler, W, Doyle, L.A, Park, H, Bick, M.J, Mao, B, Foight, G.W, Lee, M, Carter, L, Sankaran, B, Ovchinnikov, S, Marcos, E, Huang, P, Vaughan, J.C, Stoddard, B.L, Baker, D. | | Deposit date: | 2018-04-10 | | Release date: | 2018-09-19 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | De novo design of a fluorescence-activating beta-barrel.
Nature, 561, 2018
|
|
6NUK
 
 | | De novo designed protein Ferredog-Diesel | | Descriptor: | Ferredog-Diesel | | Authors: | Koepnick, B, Bick, M.J, DiMaio, F, Norgard-Solano, T, Baker, D. | | Deposit date: | 2019-02-01 | | Release date: | 2019-06-12 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | De novo protein design by citizen scientists.
Nature, 570, 2019
|
|
6NY8
 
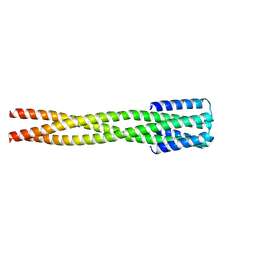 | |
6NYE
 
 | |
6NZ1
 
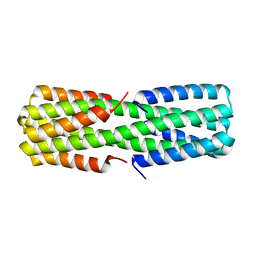 | |
6NXM
 
 | |
6NX2
 
 | | Crystal structure of computationally designed protein AAA | | Descriptor: | BROMIDE ION, Design construct AAA | | Authors: | Wei, K.Y, Bick, M.J. | | Deposit date: | 2019-02-07 | | Release date: | 2020-04-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Computational design of closely related proteins that adopt two well-defined but structurally divergent folds.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6NYI
 
 | |
6NYK
 
 | |
6NZ3
 
 | |
7LZ3
 
 | | Computational design of constitutively active cGAS | | Descriptor: | Cyclic GMP-AMP synthase, GLYCEROL, ZINC ION | | Authors: | Dowling, Q, Volkman, H.E, Gray, E.E, Ovchinnikov, S, Cambier, S, Bera, A.K, Bick, M, Kang, A, Stetson, D.B, King, N.P. | | Deposit date: | 2021-03-08 | | Release date: | 2022-03-16 | | Last modified: | 2023-02-01 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Computational design of constitutively active cGAS.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8SZZ
 
 | |
6TMS
 
 | | Crystal structure of a de novo designed hexameric helical-bundle protein | | Descriptor: | SULFATE ION, a novel designed pore protein, affinity purification tag | | Authors: | Xu, C, Pei, X.Y, Luisi, B.F, Baker, D. | | Deposit date: | 2019-12-05 | | Release date: | 2020-04-29 | | Last modified: | 2020-09-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Computational design of transmembrane pores.
Nature, 585, 2020
|
|
6TJ1
 
 | | Crystal structure of a de novo designed hexameric helical-bundle protein | | Descriptor: | De novo designed WSHC6, purification tag | | Authors: | Xu, C, Pei, X.Y, Luisi, B.F, Baker, D. | | Deposit date: | 2019-11-23 | | Release date: | 2020-04-29 | | Last modified: | 2020-09-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Computational design of transmembrane pores.
Nature, 585, 2020
|
|
