1IIY
 
 | |
2EZM
 
 | |
2EZN
 
 | |
2Y1S
 
 | | Microvirin lectin | | Descriptor: | Mannan-binding lectin | | Authors: | Bewley, C.A, Hussan, S. | | Deposit date: | 2010-12-10 | | Release date: | 2011-04-06 | | Last modified: | 2024-10-23 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the monovalent lectin microvirin in complex with Man(alpha)(1-2)Man provides a basis for anti-HIV activity with low toxicity.
J. Biol. Chem., 286, 2011
|
|
7MGV
 
 | | Chryseobacterium gregarium RiPP-associated ATP-grasp ligase in complex with ADP, and a leader and core peptide | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CdnA3 Core peptide, CdnA3 Leader peptide, ... | | Authors: | Bewley, C.A, Zhao, G, Kosek, D, Dyda, F. | | Deposit date: | 2021-04-13 | | Release date: | 2021-06-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Structural Basis for a Dual Function ATP Grasp Ligase That Installs Single and Bicyclic omega-Ester Macrocycles in a New Multicore RiPP Natural Product.
J.Am.Chem.Soc., 143, 2021
|
|
2RLL
 
 | | CCR5 Nt(7-15) | | Descriptor: | 9-mer from C-C chemokine receptor type 5 | | Authors: | Bewley, C.A, Lam, S.N. | | Deposit date: | 2007-07-21 | | Release date: | 2007-09-25 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Structures of the CCR5 N terminus and of a tyrosine-sulfated antibody with HIV-1 gp120 and CD4
Science, 317, 2007
|
|
2YHH
 
 | | Microvirin:mannobiose complex | | Descriptor: | MANNAN-BINDING LECTIN, alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose | | Authors: | Hussan, S, Bewley, C.A, Clore, G.M, Gustchina, E, Ghirlando, R. | | Deposit date: | 2011-05-02 | | Release date: | 2011-06-15 | | Last modified: | 2020-07-29 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the Monovalent Lectin Microvirin in Complex with Man(Alpha)(1-2)Man Provides a Basis for Anti-HIV Activity with Low Toxicity.
J.Biol.Chem., 286, 2011
|
|
1J4V
 
 | | CYANOVIRIN-N | | Descriptor: | CYANOVIRIN-N | | Authors: | Clore, G.M, Bewley, C.A. | | Deposit date: | 2001-11-21 | | Release date: | 2002-03-06 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Using conjoined rigid body/torsion angle simulated annealing to determine the relative orientation of covalently linked protein domains from dipolar couplings.
J.Magn.Reson., 154, 2002
|
|
7RIC
 
 | | Griffithsin variant Y28W/Y68W/Y110W | | Descriptor: | Griffithsin | | Authors: | Zhao, G, Sun, J, Bewley, C.A. | | Deposit date: | 2021-07-19 | | Release date: | 2022-05-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | C 3 -Symmetric Aromatic Core of Griffithsin Is Essential for Potent Anti-HIV Activity.
Acs Chem.Biol., 17, 2022
|
|
7RID
 
 | | Griffithsin variant Y28A | | Descriptor: | Griffithsin | | Authors: | Zhao, G, Sun, J, Bewley, C.A. | | Deposit date: | 2021-07-19 | | Release date: | 2022-05-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | C 3 -Symmetric Aromatic Core of Griffithsin Is Essential for Potent Anti-HIV Activity.
Acs Chem.Biol., 17, 2022
|
|
7RIB
 
 | | Griffithsin mutant Y28F/Y68F/Y110F | | Descriptor: | Griffithsin, SULFATE ION, alpha-D-mannopyranose | | Authors: | Zhao, G, Sun, J, Bewley, C.A. | | Deposit date: | 2021-07-19 | | Release date: | 2022-05-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | C 3 -Symmetric Aromatic Core of Griffithsin Is Essential for Potent Anti-HIV Activity.
Acs Chem.Biol., 17, 2022
|
|
7RIA
 
 | | Griffithsin variant Y28A/Y68A/Y110A | | Descriptor: | Griffithsin, SULFATE ION, alpha-D-mannopyranose | | Authors: | Zhao, G, Sun, J, Bewley, C.A. | | Deposit date: | 2021-07-19 | | Release date: | 2022-05-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | C 3 -Symmetric Aromatic Core of Griffithsin Is Essential for Potent Anti-HIV Activity.
Acs Chem.Biol., 17, 2022
|
|
1ZHS
 
 | | Crystal structure of MVL bound to Man3GlcNAc2 | | Descriptor: | 1,2-ETHANEDIOL, PHOSPHATE ION, alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Williams, D.C, Lee, J.Y, Cai, M, Bewley, C.A, Clore, G.M. | | Deposit date: | 2005-04-26 | | Release date: | 2005-06-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structures of the HIV-1 Inhibitory Cyanobacterial Protein MVL Free and Bound to Man3GlcNAc2: STRUCTURAL BASIS FOR SPECIFICITY AND HIGH-AFFINITY BINDING TO THE CORE PENTASACCHARIDE FROM N-LINKED OLIGOMANNOSIDE.
J.Biol.Chem., 280, 2005
|
|
1ZHQ
 
 | | Crystal structure of apo MVL | | Descriptor: | 1,2-ETHANEDIOL, PHOSPHATE ION, mannan-binding lectin | | Authors: | Williams, D.C, Lee, J.Y, Cai, M, Bewley, C.A, Clore, G.M. | | Deposit date: | 2005-04-26 | | Release date: | 2005-06-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structures of the HIV-1 Inhibitory Cyanobacterial Protein MVL Free and Bound to Man3GlcNAc2: STRUCTURAL BASIS FOR SPECIFICITY AND HIGH-AFFINITY BINDING TO THE CORE PENTASACCHARIDE FROM N-LINKED OLIGOMANNOSIDE.
J.Biol.Chem., 280, 2005
|
|
6BF4
 
 | |
2QAD
 
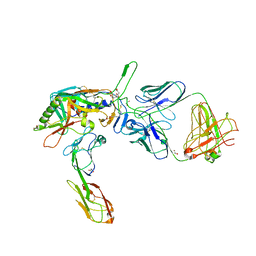 | | Structure of tyrosine-sulfated 412d antibody complexed with HIV-1 YU2 gp120 and CD4 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein gp160, ... | | Authors: | Huang, C.-C, Tang, M, Robinson, J, Wyatt, R, Kwong, P.D. | | Deposit date: | 2007-06-14 | | Release date: | 2007-09-25 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structures of the CCR5 N terminus and of a tyrosine-sulfated antibody with HIV-1 gp120 and CD4
Science, 317, 2007
|
|
5FYJ
 
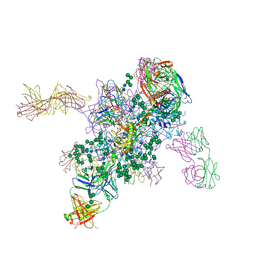 | | Crystal Structure at 3.4 A Resolution of Fully Glycosylated HIV-1 Clade G X1193.c1 SOSIP.664 Prefusion Env Trimer in Complex with Broadly Neutralizing Antibodies PGT122, 35O22 and VRC01 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Stewart-Jones, G.B.E, Zhou, T, Thomas, P.V, Kwong, P.D. | | Deposit date: | 2016-03-08 | | Release date: | 2016-04-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.11 Å) | | Cite: | Trimeric HIV-1-Env Structures Define Glycan Shields from Clades A, B and G
Cell(Cambridge,Mass.), 165, 2016
|
|
5FYK
 
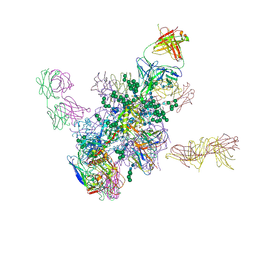 | | Crystal Structure at 3.7 A Resolution of Fully Glycosylated HIV-1 Clade B JR-FL SOSIP.664 Prefusion Env Trimer in Complex with Broadly Neutralizing Antibodies PGT122, 35O22 and VRC01 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 35O22, ... | | Authors: | Stewart-Jones, G.B.E, Zhou, T, Thomas, P.V, Kwong, P.D. | | Deposit date: | 2016-03-08 | | Release date: | 2016-04-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.107 Å) | | Cite: | Trimeric HIV-1-Env Structures Define Glycan Shields from Clades A, B and G
Cell(Cambridge,Mass.), 165, 2016
|
|
5FYL
 
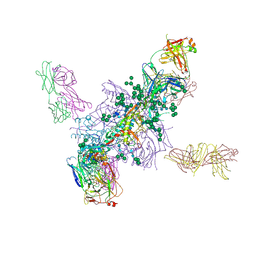 | | Crystal Structure at 3.7 A Resolution of Fully Glycosylated HIV-1 Clade A BG505 SOSIP.664 Prefusion Env Trimer in Complex with Broadly Neutralizing Antibodies PGT122 and 35O22 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 35O22 ANTIBODY FAB HEAVY CHAIN, ... | | Authors: | Stewart-Jones, G.B.E, Zhou, T, Thomas, P.V, Kwong, P.D. | | Deposit date: | 2016-03-08 | | Release date: | 2016-04-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Trimeric HIV-1-Env Structures Define Glycan Shields from Clades A, B and G
Cell(Cambridge,Mass.), 165, 2016
|
|
7RKG
 
 | | Griffithsin mutant Y28W | | Descriptor: | Griffithsin, alpha-D-mannopyranose | | Authors: | Zhao, G, Sun, J, Bewley, C. | | Deposit date: | 2021-07-22 | | Release date: | 2022-05-25 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | C 3 -Symmetric Aromatic Core of Griffithsin Is Essential for Potent Anti-HIV Activity.
Acs Chem.Biol., 17, 2022
|
|
2EZD
 
 | | SOLUTION STRUCTURE OF A COMPLEX OF THE SECOND DNA BINDING DOMAIN OF HUMAN HMG-I(Y) BOUND TO DNA DODECAMER CONTAINING THE PRDII SITE OF THE INTERFERON-BETA PROMOTER, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | DNA (5'-D(*GP*AP*GP*GP*AP*AP*TP*TP*TP*CP*CP*C)-3'), DNA (5'-D(*GP*GP*GP*AP*AP*AP*TP*TP*CP*CP*TP*C)-3'), HIGH MOBILITY GROUP PROTEIN HMG-I/HMG-Y | | Authors: | Clore, G.M, Huth, J.R, Bewley, C, Gronenborn, A.M. | | Deposit date: | 1997-06-04 | | Release date: | 1997-10-15 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The solution structure of an HMG-I(Y)-DNA complex defines a new architectural minor groove binding motif.
Nat.Struct.Biol., 4, 1997
|
|
2EZE
 
 | | SOLUTION STRUCTURE OF A COMPLEX OF THE SECOND DNA BINDING DOMAIN OF HUMAN HMG-I(Y) BOUND TO DNA DODECAMER CONTAINING THE PRDII SITE OF THE INTERFERON-BETA PROMOTER, NMR, 35 STRUCTURES | | Descriptor: | DNA (5'-D(*GP*AP*GP*GP*AP*AP*TP*TP*TP*CP*CP*C)-3'), DNA (5'-D(*GP*GP*GP*AP*AP*AP*TP*TP*CP*CP*TP*C)-3'), HIGH MOBILITY GROUP PROTEIN HMG-I/HMG-Y | | Authors: | Clore, G.M, Huth, J.R, Bewley, C, Gronenborn, A.M. | | Deposit date: | 1997-06-04 | | Release date: | 1997-10-15 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | The solution structure of an HMG-I(Y)-DNA complex defines a new architectural minor groove binding motif.
Nat.Struct.Biol., 4, 1997
|
|
2EZF
 
 | | SOLUTION STRUCTURE OF A COMPLEX OF THE THIRD DNA BINDING DOMAIN OF HUMAN HMG-I(Y) BOUND TO DNA DODECAMER CONTAINING THE PRDII SITE OF THE INTERFERON-BETA PROMOTER, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | DNA (5'-D(*GP*AP*GP*GP*AP*AP*TP*TP*TP*CP*CP*C)-3'), DNA (5'-D(*GP*GP*GP*AP*AP*AP*TP*TP*CP*CP*TP*C)-3'), HIGH MOBILITY GROUP PROTEIN HMG-I/HMG-Y | | Authors: | Clore, G.M, Huth, J.R, Bewley, C, Gronenborn, A.M. | | Deposit date: | 1997-06-04 | | Release date: | 1997-10-15 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | The solution structure of an HMG-I(Y)-DNA complex defines a new architectural minor groove binding motif.
Nat.Struct.Biol., 4, 1997
|
|
2EZG
 
 | | SOLUTION STRUCTURE OF A COMPLEX OF THE THIRD DNA BINDING DOMAIN OF HUMAN HMG-I(Y) BOUND TO DNA DODECAMER CONTAINING THE PRDII SITE OF THE INTERFERON-BETA PROMOTER, NMR, 35 STRUCTURES | | Descriptor: | DNA (5'-D(*GP*AP*GP*GP*AP*AP*TP*TP*TP*CP*CP*C)-3'), DNA (5'-D(*GP*GP*GP*AP*AP*AP*TP*TP*CP*CP*TP*C)-3'), HIGH MOBILITY GROUP PROTEIN HMG-I/HMG-Y | | Authors: | Clore, G.M, Huth, J.R, Bewley, C, Gronenborn, A.M. | | Deposit date: | 1997-06-04 | | Release date: | 1997-10-15 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | The solution structure of an HMG-I(Y)-DNA complex defines a new architectural minor groove binding motif.
Nat.Struct.Biol., 4, 1997
|
|
3RY8
 
 | | Structural basis for norovirus inhibition and fucose mimicry by citrate | | Descriptor: | 1,2-ETHANEDIOL, CITRATE ANION, Capsid protein | | Authors: | Hansman, G.S, McLellan, J.S, Kwong, P.D. | | Deposit date: | 2011-05-11 | | Release date: | 2011-11-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural basis for norovirus inhibition and fucose mimicry by citrate.
J.Virol., 86, 2012
|
|
