4TN9
 
 | | Crystal structure of Clostridium histolyticum ColG collagenase polycystic kidney disease-like domain at 1.4 Angstrom resolution | | Descriptor: | Collagenase | | Authors: | Bauer, R, Sakon, J, Philominathan, S.T.L, Matsushita, O, Gann, S. | | Deposit date: | 2014-06-03 | | Release date: | 2015-03-11 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structures of three polycystic kidney disease-like domains from Clostridium histolyticum collagenases ColG and ColH.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4U6T
 
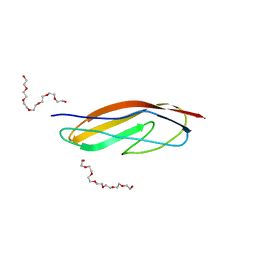 | | Crystal structure of the Clostridium histolyticum colH collagenase polycystic kidney disease-like domain 2a at 1.76 Angstrom resolution | | Descriptor: | 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, ColH protein | | Authors: | Bauer, R, Janowska, K, Sakon, J, Matsushita, O, Latimer, E. | | Deposit date: | 2014-07-29 | | Release date: | 2015-03-11 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Structures of three polycystic kidney disease-like domains from Clostridium histolyticum collagenases ColG and ColH.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4U7K
 
 | | Crystal structure of the Clostridium histolyticum colH collagenase polycystic kidney disease-like domain 2a in the presence of calcium at 1.9 Angstrom resolution | | Descriptor: | CALCIUM ION, ColH protein | | Authors: | Bauer, R, Janowska, K, Sakon, J, Matsushita, O, Latimer, E. | | Deposit date: | 2014-07-31 | | Release date: | 2015-03-11 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Structures of three polycystic kidney disease-like domains from Clostridium histolyticum collagenases ColG and ColH.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4JGU
 
 | | Crystal structure of Clostridium histolyticum ColH collagenase polycystic kidney-disease-like domain 2b at 1.4 Angstrom resolution in the presence of calcium | | Descriptor: | CALCIUM ION, ColH protein, SULFATE ION | | Authors: | Bauer, R, Matsushita, O, Sakon, J. | | Deposit date: | 2013-03-03 | | Release date: | 2014-03-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Structures of three polycystic kidney disease-like domains from Clostridium histolyticum collagenases ColG and ColH.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
1VET
 
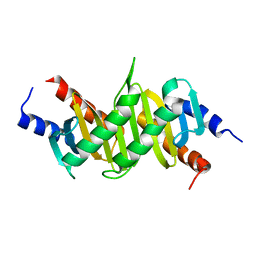 | | Crystal Structure of p14/MP1 at 1.9 A resolution | | Descriptor: | Late endosomal/lysosomal Mp1 interacting protein, Mitogen-activated protein kinase kinase 1 interacting protein 1 | | Authors: | Kurzbauer, R, Teis, D, Maurer-Stroh, S, Eisenhaber, F, Hekman, M, Bourenkov, G.P, Bartunik, H.D, Huber, L.A, Clausen, T. | | Deposit date: | 2004-04-05 | | Release date: | 2004-08-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the p14/MP1 scaffolding complex: How a twin couple attaches mitogen- activated protein kinase signaling to late endosomes
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
1VEU
 
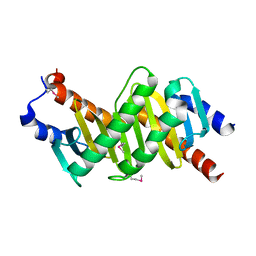 | | Crystal structure of the p14/MP1 complex at 2.15 A resolution | | Descriptor: | Late endosomal/lysosomal Mp1 interacting protein, Mitogen-activated protein kinase kinase 1 interacting protein 1 | | Authors: | Kurzbauer, R, Teis, D, Maurer-Stroh, S, Eisenhaber, F, Hekman, M, Bourenkov, G.P, Bartunik, H.D, Huber, L.A, Clausen, T. | | Deposit date: | 2004-04-05 | | Release date: | 2004-08-03 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structure of the p14/MP1 scaffolding complex: How a twin couple attaches mitogen- activated protein kinase signaling to late endosomes
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
6MPX
 
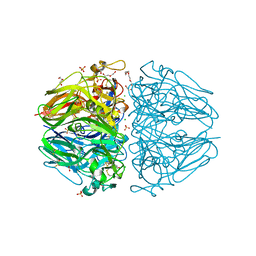 | | Twelve chloride ions induce formation and stabilize the NC1 hexamer of collagen IV assembled from transition state trimers | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Bauer, R, Boudko, S.P, Hudson, B.G. | | Deposit date: | 2018-10-09 | | Release date: | 2019-04-03 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A chloride ring is an ancient evolutionary innovation mediating the assembly of the collagen IV scaffold of basement membranes.
J.Biol.Chem., 294, 2019
|
|
3JQW
 
 | | Crystal structure of Clostridium histolyticum colH collagenase collagen-binding domain 3 at 2 Angstrom resolution in presence of calcium | | Descriptor: | CALCIUM ION, ColH protein | | Authors: | Sakon, J, Philominathan, S.T.L, Matsushita, O, Bauer, R. | | Deposit date: | 2009-09-08 | | Release date: | 2010-09-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Comparison of ColH and ColG Collagen-Binding Domains from Clostridium histolyticum.
J.Bacteriol., 195, 2013
|
|
3JQX
 
 | | Crystal structure of Clostridium histolyticum colH collagenase collagen binding domain 3 at 2.2 Angstrom resolution in the presence of calcium and cadmium | | Descriptor: | CADMIUM ION, CALCIUM ION, ColH protein | | Authors: | Sakon, J, Philominathan, S.T.L, Bauer, R, Matsushita, O. | | Deposit date: | 2009-09-08 | | Release date: | 2010-09-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Comparison of ColH and ColG Collagen-Binding Domains from Clostridium histolyticum.
J.Bacteriol., 195, 2013
|
|
4HPK
 
 | | Crystal structure of Clostridium histolyticum colg collagenase collagen-binding domain 3B at 1.35 Angstrom resolution in presence of calcium nitrate | | Descriptor: | CALCIUM ION, CHLORIDE ION, Collagenase, ... | | Authors: | Philominathan, S.T.L, Wilson, J.J, Bauer, R, Matsushita, O, Sakon, J. | | Deposit date: | 2012-10-24 | | Release date: | 2012-12-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structural Comparison of ColH and ColG Collagen-Binding Domains from Clostridium histolyticum.
J.Bacteriol., 195, 2013
|
|
4JRW
 
 | | Crystal structure of Clostridium histolyticum colg collagenase PKD domain 2 at 1.6 Angstrom resolution | | Descriptor: | BROMIDE ION, Collagenase | | Authors: | Sakon, J, Philominathan, S.T.L, Gann, S, Bauer, R, Matsushita, O. | | Deposit date: | 2013-03-22 | | Release date: | 2014-05-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structures of three polycystic kidney disease-like domains from Clostridium histolyticum collagenases ColG and ColH.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
5IKU
 
 | | Crystal structure of the Hathewaya histolytica ColG tandem collagen-binding domain s3as3b in the presence of calcium at 1.9 Angstrom resolution | | Descriptor: | CALCIUM ION, Collagenase | | Authors: | Janowska, K, Bauer, R, Roeser, R, Sakon, J, Matsushita, O. | | Deposit date: | 2016-03-03 | | Release date: | 2017-03-15 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Ca2+-induced orientation of tandem collagen binding domains from clostridial collagenase ColG permits two opposing functions of collagen fibril formation and retardation.
Febs J., 285, 2018
|
|
1GG1
 
 | | CRYSTAL STRUCTURE ANALYSIS OF DAHP SYNTHASE IN COMPLEX WITH MN2+ AND 2-PHOSPHOGLYCOLATE | | Descriptor: | 2-PHOSPHOGLYCOLIC ACID, 3-DEOXY-D-ARABINO-HEPTULOSONATE-7-PHOSPHATE SYNTHASE, MANGANESE (II) ION, ... | | Authors: | Wagner, T, Shumilin, I.A, Bauerle, R, Kretsinger, R.H. | | Deposit date: | 2000-08-04 | | Release date: | 2000-10-04 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of 3-deoxy-d-arabino-heptulosonate-7-phosphate synthase from Escherichia coli: comparison of the Mn(2+)*2-phosphoglycolate and the Pb(2+)*2-phosphoenolpyruvate complexes and implications for catalysis.
J.Mol.Biol., 301, 2000
|
|
3JUO
 
 | | Crystal Structure of PhzA/B from Burkholderia cepacia R18194 in complex with (R)-5-bromo-2-(piperidin-3-ylamino)benzoic acid | | Descriptor: | 5-bromo-2-[(3R)-piperidin-3-ylamino]benzoic acid, Phenazine biosynthesis protein A/B | | Authors: | Mentel, M, Jain, I.H, Breinbauer, R, Blankenfeldt, W. | | Deposit date: | 2009-09-15 | | Release date: | 2009-09-29 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Active Site of an Enzyme Can Host Both Enantiomers of a Racemic Ligand Simultaneously
Angew.Chem.Int.Ed.Engl., 48, 2009
|
|
3JUP
 
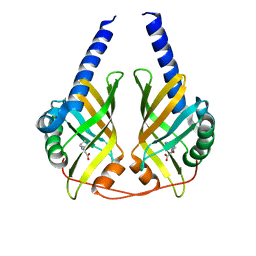 | | Crystal Structure of PhzA/B from Burkholderia cepacia R18194 in complex with (S)-5-bromo-2-(piperidin-3-ylamino)benzoic acid | | Descriptor: | 5-bromo-2-[(3S)-piperidin-3-ylamino]benzoate, Phenazine biosynthesis protein A/B | | Authors: | Mentel, M, Jain, I.H, Breinbauer, R, Blankenfeldt, W. | | Deposit date: | 2009-09-15 | | Release date: | 2009-09-29 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Active Site of an Enzyme Can Host Both Enantiomers of a Racemic Ligand Simultaneously
Angew.Chem.Int.Ed.Engl., 48, 2009
|
|
3JUN
 
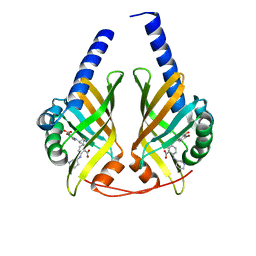 | |
3JUM
 
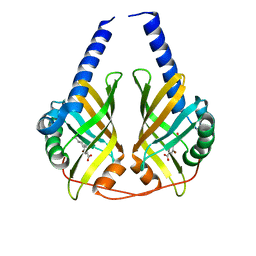 | | Crystal Structure of PhzA/B from Burkholderia cepacia R18194 in complex with 5-bromo-2-((1S,3R)-3-carboxycyclohexylamino)benzoic acid | | Descriptor: | 5-bromo-2-{[(1S,3R)-3-carboxycyclohexyl]amino}benzoic acid, Phenazine biosynthesis protein A/B | | Authors: | Mentel, M, Breinbauer, R, Blankenfeldt, W. | | Deposit date: | 2009-09-15 | | Release date: | 2009-09-29 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | The Active Site of an Enzyme Can Host Both Enantiomers of a Racemic Ligand Simultaneously
Angew.Chem.Int.Ed.Engl., 48, 2009
|
|
3JUQ
 
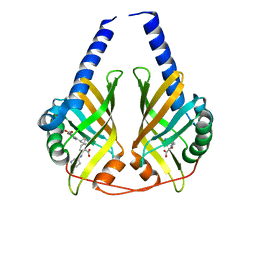 | |
1GG0
 
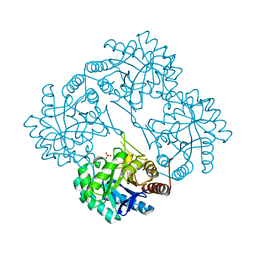 | | CRYSTAL STRUCTURE ANALYSIS OF KDOP SYNTHASE AT 3.0 A | | Descriptor: | 3-DEOXY-D-MANNO-OCTULOSONATE 8-PHOSPHATE SYNTHASE, PHOSPHATE ION | | Authors: | Wagner, T, Kretsinger, R.H, Bauerle, R, Tolbert, W.D. | | Deposit date: | 2000-08-04 | | Release date: | 2000-10-04 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | 3-Deoxy-D-manno-octulosonate-8-phosphate synthase from Escherichia coli. Model of binding of phosphoenolpyruvate and D-arabinose-5-phosphate.
J.Mol.Biol., 301, 2000
|
|
4C2C
 
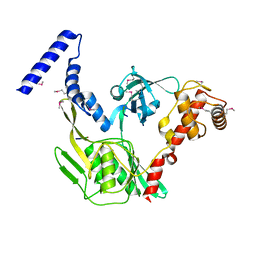 | | Crystal structure of the protease CtpB in an active state | | Descriptor: | CARBOXY-TERMINAL PROCESSING PROTEASE CTPB, PEPTIDE1, PEPTIDE2 | | Authors: | Mastny, M, Heuck, A, Kurzbauer, R, Clausen, T. | | Deposit date: | 2013-08-17 | | Release date: | 2013-12-04 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Ctpb Assembles a Gated Protease Tunnel Regulating Cell-Cell Signaling During Spore Formation in Bacillus Subtilis.
Cell(Cambridge,Mass.), 155, 2013
|
|
4C2D
 
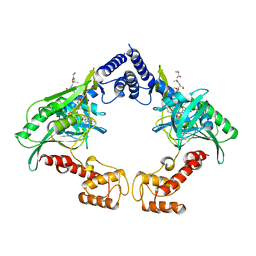 | | Crystal structure of the protease CtpB in an active state | | Descriptor: | CARBOXY-TERMINAL PROCESSING PROTEASE CTPB, PEPTIDE1, PEPTIDE2 | | Authors: | Mastny, M, Heuck, A, Kurzbauer, R, Clausen, T. | | Deposit date: | 2013-08-17 | | Release date: | 2013-12-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Ctpb Assembles a Gated Protease Tunnel Regulating Cell-Cell Signaling During Spore Formation in Bacillus Subtilis.
Cell(Cambridge,Mass.), 155, 2013
|
|
4C2E
 
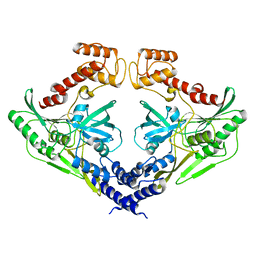 | | Crystal structure of the protease CtpB(S309A) present in a resting state | | Descriptor: | CARBOXY-TERMINAL PROCESSING PROTEASE CTPB | | Authors: | Mastny, M, Heuck, A, Kurzbauer, R, Clausen, T. | | Deposit date: | 2013-08-17 | | Release date: | 2013-12-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Ctpb Assembles a Gated Protease Tunnel Regulating Cell-Cell Signaling During Spore Formation in Bacillus Subtilis.
Cell(Cambridge,Mass.), 155, 2013
|
|
4C2G
 
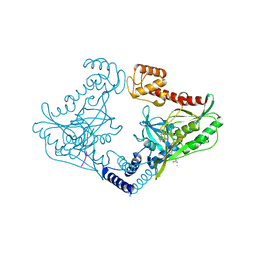 | | Crystal structure of CtpB(S309A) in complex with a peptide having a Val-Pro-Ala C-terminus | | Descriptor: | CARBOXY-TERMINAL PROCESSING PROTEASE CTPB, PEPTIDE1 | | Authors: | Mastny, M, Heuck, A, Kurzbauer, R, Clausen, T. | | Deposit date: | 2013-08-17 | | Release date: | 2013-12-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Ctpb Assembles a Gated Protease Tunnel Regulating Cell-Cell Signaling During Spore Formation in Bacillus Subtilis.
Cell(Cambridge,Mass.), 155, 2013
|
|
4C2H
 
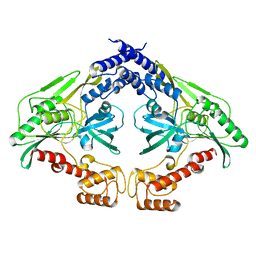 | | Crystal structure of the CtpB(V118Y) mutant | | Descriptor: | CARBOXY-TERMINAL PROCESSING PROTEASE CTPB | | Authors: | Mastny, M, Heuck, A, Kurzbauer, R, Clausen, T. | | Deposit date: | 2013-08-17 | | Release date: | 2013-12-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Ctpb Assembles a Gated Protease Tunnel Regulating Cell-Cell Signaling During Spore Formation in Bacillus Subtilis.
Cell(Cambridge,Mass.), 155, 2013
|
|
4C2F
 
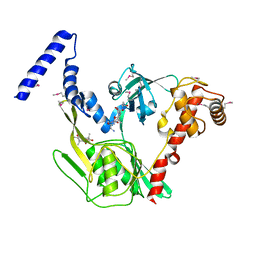 | | Crystal structure of the CtpB R168A mutant present in an active conformation | | Descriptor: | CARBOXY-TERMINAL PROCESSING PROTEASE CTPB, PEPTIDE1, PEPTIDE2 | | Authors: | Mastny, M, Heuck, A, Kurzbauer, R, Clausen, T. | | Deposit date: | 2013-08-17 | | Release date: | 2013-12-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Ctpb Assembles a Gated Protease Tunnel Regulating Cell-Cell Signaling During Spore Formation in Bacillus Subtilis.
Cell(Cambridge,Mass.), 155, 2013
|
|
