3RIX
 
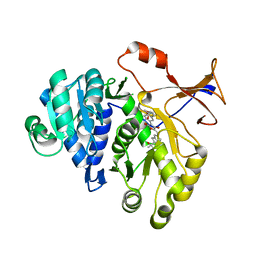 | | 1.7A resolution structure of a firefly luciferase-Aspulvinone J inhibitor complex | | Descriptor: | (5Z)-4-hydroxy-3-[(2R)-2-(2-hydroxypropan-2-yl)-2,3-dihydro-1-benzofuran-5-yl]-5-{[(2R)-2-(2-hydroxypropan-2-yl)-2,3-dihydro-1-benzofuran-5-yl]methylidene}furan-2(5H)-one, Luciferin 4-monooxygenase | | Authors: | Lovell, S, Battaile, K.P, Lopez, P.C, Auld, D.S, Schultz, P.J, MacArthur, R, Shen, M, Tamayo, G, Inglese, J, Sherman, D.H. | | Deposit date: | 2011-04-14 | | Release date: | 2011-12-07 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Titration-based screening for evaluation of natural product extracts: identification of an aspulvinone family of luciferase inhibitors.
Chem.Biol., 18, 2011
|
|
3R2L
 
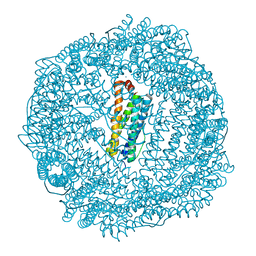 | | 1.85A resolution structure of Iron Soaked FtnA from Pseudomonas aeruginosa (pH 7.5) | | Descriptor: | Bacterioferritin, FE (III) ION, SODIUM ION | | Authors: | Lovell, S.W, Battaile, K.P, Yao, H, Jepkorir, G, Nama, P.V, Weeratunga, S, Rivera, M. | | Deposit date: | 2011-03-14 | | Release date: | 2011-05-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Two distinct ferritin-like molecules in Pseudomonas aeruginosa: the product of the bfrA gene is a bacterial ferritin (FtnA) and not a bacterioferritin (Bfr).
Biochemistry, 50, 2011
|
|
3R2S
 
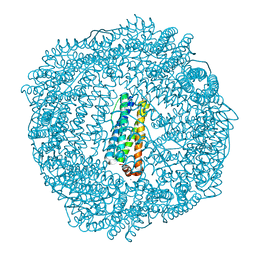 | | 2.1A resolution structure of Doubly Soaked FtnA from Pseudomonas aeruginosa (pH 6.0) | | Descriptor: | Bacterioferritin, FE (III) ION, SODIUM ION, ... | | Authors: | Lovell, S.W, Battaile, K.P, Yao, H, Jepkorir, G, Nama, P.V, Weeratunga, S, Rivera, M. | | Deposit date: | 2011-03-14 | | Release date: | 2011-05-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Two distinct ferritin-like molecules in Pseudomonas aeruginosa: the product of the bfrA gene is a bacterial ferritin (FtnA) and not a bacterioferritin (Bfr).
Biochemistry, 50, 2011
|
|
3R9J
 
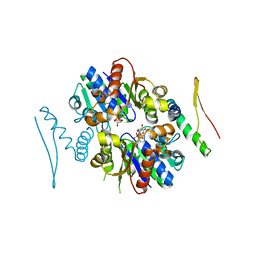 | | 4.3A resolution structure of a MinD-MinE(I24N) protein complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Cell division topological specificity factor, Septum site-determining protein minD | | Authors: | Lovell, S, Battaile, K.P, Park, K.-T, Wu, W, Holyoak, T, Lutkenhaus, J. | | Deposit date: | 2011-03-25 | | Release date: | 2011-08-17 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (4.3 Å) | | Cite: | The Min Oscillator Uses MinD-Dependent Conformational Changes in MinE to Spatially Regulate Cytokinesis.
Cell(Cambridge,Mass.), 146, 2011
|
|
3R2R
 
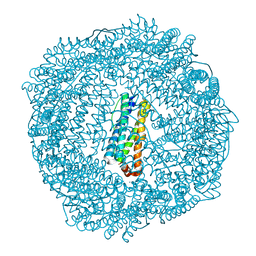 | | 1.65A resolution structure of Iron Soaked FtnA from Pseudomonas aeruginosa (pH 6.0) | | Descriptor: | Bacterioferritin, FE (III) ION, SODIUM ION, ... | | Authors: | Lovell, S.W, Battaile, K.P, Yao, H, Jepkorir, G, Nama, P.V, Weeratunga, S, Rivera, M. | | Deposit date: | 2011-03-14 | | Release date: | 2011-05-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Two distinct ferritin-like molecules in Pseudomonas aeruginosa: the product of the bfrA gene is a bacterial ferritin (FtnA) and not a bacterioferritin (Bfr).
Biochemistry, 50, 2011
|
|
3R2M
 
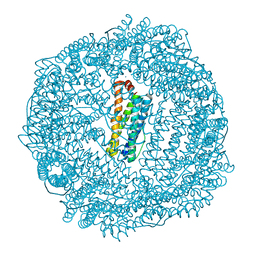 | | 1.8A resolution structure of Doubly Soaked FtnA from Pseudomonas aeruginosa (pH 7.5) | | Descriptor: | Bacterioferritin, FE (III) ION, SODIUM ION | | Authors: | Lovell, S.W, Battaile, K.P, Yao, H, Jepkorir, G, Nama, P.V, Weeratunga, S, Rivera, M. | | Deposit date: | 2011-03-14 | | Release date: | 2011-05-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Two distinct ferritin-like molecules in Pseudomonas aeruginosa: the product of the bfrA gene is a bacterial ferritin (FtnA) and not a bacterioferritin (Bfr).
Biochemistry, 50, 2011
|
|
8E7A
 
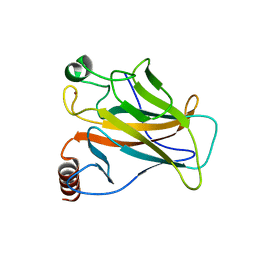 | | Crystal structure of the p53 (Y107H) core domain orthorhombic P form | | Descriptor: | Cellular tumor antigen p53, ZINC ION | | Authors: | Lovell, S, Liu, L, Battaile, K.P, Miller, S, Karanicolas, J. | | Deposit date: | 2022-08-23 | | Release date: | 2023-05-17 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | An African-Specific Variant of TP53 Reveals PADI4 as a Regulator of p53-Mediated Tumor Suppression.
Cancer Discov, 13, 2023
|
|
8E7B
 
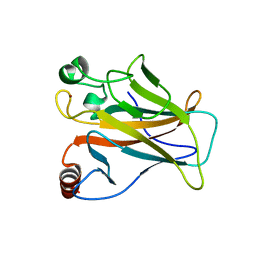 | | Crystal structure of the p53 (Y107H) core domain monoclinic P form | | Descriptor: | Cellular tumor antigen p53, ZINC ION | | Authors: | Lovell, S, Liu, L, Battaile, K.P, Miller, S, Karanicolas, J. | | Deposit date: | 2022-08-23 | | Release date: | 2023-05-17 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | An African-Specific Variant of TP53 Reveals PADI4 as a Regulator of p53-Mediated Tumor Suppression.
Cancer Discov, 13, 2023
|
|
8EJX
 
 | |
8F44
 
 | | Crystal structure of SARS-CoV-2 3CL protease in complex with a dimethyl phenyl sulfane inhibitor | | Descriptor: | (1R,2S)-1-hydroxy-2-[(N-{[2-methyl-2-(phenylsulfanyl)propoxy]carbonyl}-L-leucyl)amino]-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, (2-methyl-2-phenylsulfanyl-propyl) ~{N}-[(2~{S})-1-[[(1~{S},2~{S})-1-[bis(oxidanyl)-oxidanylidene-$l^{5}-sulfanyl]-1-oxidanyl-3-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]amino]-4-methyl-1-oxidanylidene-pentan-2-yl]carbamate, 3C-like proteinase, ... | | Authors: | Liu, L, Lovell, S, Cooper, A, Battaile, K.P, Dampalla, C.S, Groutas, W.C. | | Deposit date: | 2022-11-10 | | Release date: | 2022-11-23 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structure-guided design of direct-acting antivirals that exploit the gem-dimethyl effect and potently inhibit 3CL proteases of severe acute respiratory syndrome Coronavirus-2 (SARS-CoV-2) and middle east respiratory syndrome coronavirus (MERS-CoV)
Eur.J.Med.Chem., 254, 2023
|
|
8F45
 
 | | Crystal structure of SARS-CoV-2 3CL protease in complex with a phenyl dimethyl sulfane inhibitor (cyclopropyl ketoamide warhead) | | Descriptor: | (2-methyl-2-phenylsulfanyl-propyl) ~{N}-[(2~{S})-1-[[(2~{S},3~{S})-3-[bis(oxidanyl)-oxidanylidene-$l^{5}-sulfanyl]-4-(cyclopropylamino)-3-oxidanyl-4-oxidanylidene-1-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]butan-2-yl]amino]-4-methyl-1-oxidanylidene-pentan-2-yl]carbamate, 3C-like proteinase | | Authors: | Lovell, S, Cooper, A, Battaile, K.P, Dampalla, C.S, Groutas, W.C. | | Deposit date: | 2022-11-10 | | Release date: | 2022-11-23 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structure-guided design of direct-acting antivirals that exploit the gem-dimethyl effect and potently inhibit 3CL proteases of severe acute respiratory syndrome Coronavirus-2 (SARS-CoV-2) and middle east respiratory syndrome coronavirus (MERS-CoV)
Eur.J.Med.Chem., 254, 2023
|
|
8F46
 
 | | Crystal structure of SARS-CoV-2 3CL protease in complex with a dimethyl phenyl sulfane inhibitor (cyano warhead) | | Descriptor: | 3C-like proteinase, N-{(1Z,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-N~2~-{[2-methyl-2-(phenylsulfanyl)propoxy]carbonyl}-L-leucinamide, TETRAETHYLENE GLYCOL | | Authors: | Liu, L, Lovell, S, Cooper, A, Battaile, K.P, Dampalla, C.S, Groutas, W.C. | | Deposit date: | 2022-11-10 | | Release date: | 2022-11-23 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure-guided design of direct-acting antivirals that exploit the gem-dimethyl effect and potently inhibit 3CL proteases of severe acute respiratory syndrome Coronavirus-2 (SARS-CoV-2) and middle east respiratory syndrome coronavirus (MERS-CoV)
Eur.J.Med.Chem., 254, 2023
|
|
8FFC
 
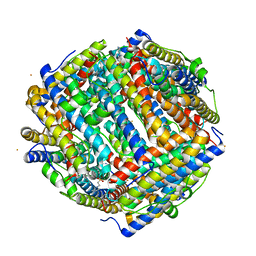 | | Crystal structure of iron bound Dps protein (PA0962) from Pseudomonas aeruginosa (cubic form) | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, FE (II) ION, Probable dna-binding stress protein | | Authors: | Lovell, S, Kashipathy, M.M, Battaile, K.P, Rivera, M. | | Deposit date: | 2022-12-08 | | Release date: | 2023-03-08 | | Last modified: | 2023-03-22 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Pseudomonas aeruginosa Dps (PA0962) Functions in H 2 O 2 Mediated Oxidative Stress Defense and Exhibits In Vitro DNA Cleaving Activity.
Int J Mol Sci, 24, 2023
|
|
8FFA
 
 | |
8FFB
 
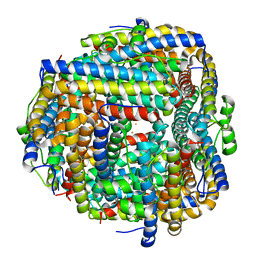 | | Crystal structure of iron bound Dps protein (PA0962) from Pseudomonas aeruginosa (orthorhombic form) | | Descriptor: | FE (II) ION, Probable dna-binding stress protein | | Authors: | Lovell, S, Kashipathy, M.M, Battaile, K.P, Rivera, M. | | Deposit date: | 2022-12-08 | | Release date: | 2023-03-08 | | Last modified: | 2023-03-22 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Pseudomonas aeruginosa Dps (PA0962) Functions in H 2 O 2 Mediated Oxidative Stress Defense and Exhibits In Vitro DNA Cleaving Activity.
Int J Mol Sci, 24, 2023
|
|
8FF9
 
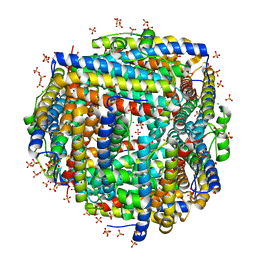 | | Crystal structure of Apo Dps protein (PA0962) from Pseudomonas aeruginosa (orthorhombic form) | | Descriptor: | CHLORIDE ION, Probable dna-binding stress protein, SODIUM ION, ... | | Authors: | Lovell, S, Kashipathy, M.M, Battaile, K.P, Rivera, M. | | Deposit date: | 2022-12-08 | | Release date: | 2023-03-08 | | Last modified: | 2023-03-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Pseudomonas aeruginosa Dps (PA0962) Functions in H 2 O 2 Mediated Oxidative Stress Defense and Exhibits In Vitro DNA Cleaving Activity.
Int J Mol Sci, 24, 2023
|
|
8FFD
 
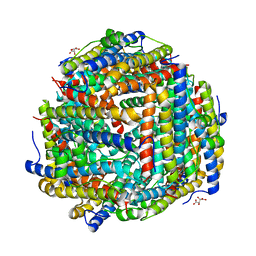 | | Crystal structure of manganeese bound Dps protein (PA0962) from Pseudomonas aeruginosa (cubic form) | | Descriptor: | L(+)-TARTARIC ACID, MANGANESE (II) ION, Probable dna-binding stress protein | | Authors: | Lovell, S, Seibold, S, Battaile, K.P, Rivera, M. | | Deposit date: | 2022-12-08 | | Release date: | 2023-03-08 | | Last modified: | 2023-03-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Pseudomonas aeruginosa Dps (PA0962) Functions in H 2 O 2 Mediated Oxidative Stress Defense and Exhibits In Vitro DNA Cleaving Activity.
Int J Mol Sci, 24, 2023
|
|
8FCH
 
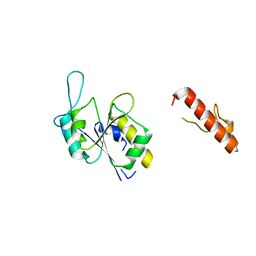 | | Apo Structure of (N1G37) Methyltransferase from Mycobacterium avium | | Descriptor: | tRNA (guanine-N(1)-)-methyltransferase | | Authors: | Isaacson, B, Brylewski, J, Schlegel, F, Cortes, J, Lavallee, T, Mehta, K, Young, A, Doti, L, Battaile, K.P, Stojanoff, V, Perez, A, Bolen, R. | | Deposit date: | 2022-12-01 | | Release date: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.956 Å) | | Cite: | Apo structure of (N1G37) Methyltransferase from Mycobacterium avium.
To Be Published
|
|
8G0G
 
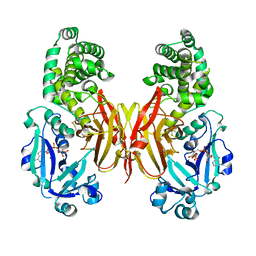 | | Crystal structure of diphtheria toxin H223Q/H257Q double mutant (pH 4.5) | | Descriptor: | ADENYLYL-3'-5'-PHOSPHO-URIDINE-3'-MONOPHOSPHATE, Diphtheria toxin | | Authors: | Lovell, S, Kashipathy, M.M, Battaile, K.P, Ladokhin, A.S. | | Deposit date: | 2023-01-31 | | Release date: | 2023-07-05 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Histidine Protonation and Conformational Switching in Diphtheria Toxin Translocation Domain.
Toxins, 15, 2023
|
|
8G0F
 
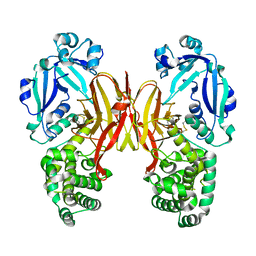 | |
7SNT
 
 | | 2.20A Resolution Structure of NanoLuc Luciferase with Bound Substrate Analog 3-methoxy-furimazine | | Descriptor: | (4S)-8-benzyl-2-[(furan-2-yl)methyl]-3-methoxy-6-phenylimidazo[1,2-a]pyrazine, Oplophorus-luciferin 2-monooxygenase catalytic subunit | | Authors: | Lovell, S, Mehzabeen, N, Battaile, K.P, Wood, M.G, Unch, J, Encell, L.P, Wood, K.V. | | Deposit date: | 2021-10-28 | | Release date: | 2022-11-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | 2.20A Resolution Structure of NanoLuc Luciferase with Bound Substrate Analog 3-methoxy-furimazine
To be published
|
|
7SNR
 
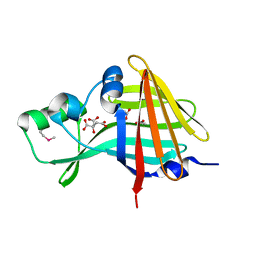 | | 2.00A Resolution Structure of NanoLuc Luciferase | | Descriptor: | ACETATE ION, CITRATE ANION, Oplophorus-luciferin 2-monooxygenase catalytic subunit | | Authors: | Lovell, S, Mehzabeen, N, Battaile, K.P, Wood, M.G, Encell, L.P, Wood, K.V. | | Deposit date: | 2021-10-28 | | Release date: | 2022-11-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | 2.00A Resolution Structure of NanoLuc Luciferase
To be published
|
|
7SNX
 
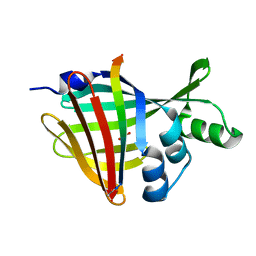 | | 1.70A Resolution Structure of NanoBiT Complementation Reporter Complex of LgBit and SmBiT Subunits | | Descriptor: | GLYCEROL, Oplophorus-luciferin 2-monooxygenase catalytic subunit, Oplophorus-luciferin 2-monooxygenase catalytic subunit: C-terminal Peptide (11-mer) | | Authors: | Lovell, S, Mehzabeen, N, Battaile, K.P, Wood, M.G, Encell, L.P, Wood, K.V. | | Deposit date: | 2021-10-28 | | Release date: | 2022-11-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | 1.70A Resolution Structure of NanoBiT Complementation Reporter Complex of LgBit and SmBiT Subunits
To be published
|
|
7SNS
 
 | | 1.55A Resolution Structure of NanoLuc Luciferase | | Descriptor: | ACETATE ION, Oplophorus-luciferin 2-monooxygenase catalytic subunit | | Authors: | Lovell, S, Mehzabeen, N, Battaile, K.P, Wood, M.G, Encell, L.P, Wood, K.V. | | Deposit date: | 2021-10-28 | | Release date: | 2022-11-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | 1.55A Resolution Structure of NanoLuc Luciferase
To be published
|
|
7SNY
 
 | | 2.10A Resolution Structure of NanoBiT Complementation Reporter Large Subunit LgBiT | | Descriptor: | Oplophorus-luciferin 2-monooxygenase catalytic subunit | | Authors: | Lovell, S, Mehzabeen, N, Battaile, K.P, Wood, M.G, Encell, L.P, Wood, K.V. | | Deposit date: | 2021-10-28 | | Release date: | 2022-11-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | 2.10A Resolution Structure of NanoBiT Complementation Reporter Large Subunit LgBiT
To be published
|
|
