1LJO
 
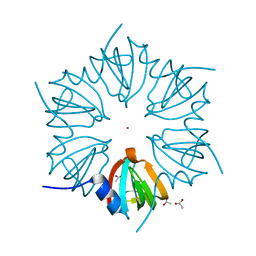 | | CRYSTAL STRUCTURE OF AN SM-LIKE PROTEIN (AF-SM2) FROM ARCHAEOGLOBUS FULGIDUS AT 1.95A RESOLUTION | | Descriptor: | ACETIC ACID, Archaeal Sm-like protein AF-Sm2, CADMIUM ION | | Authors: | Toro, I, Basquin, J, Teo-Dreher, H, Suck, D. | | Deposit date: | 2002-04-22 | | Release date: | 2002-07-03 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Archaeal Sm proteins form heptameric and hexameric complexes: crystal structures of the Sm1 and Sm2 proteins from the hyperthermophile Archaeoglobus fulgidus.
J.Mol.Biol., 320, 2002
|
|
7AQO
 
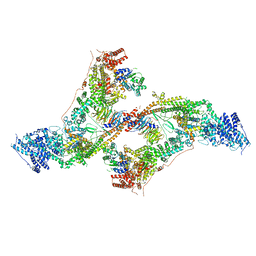 | | yeast THO-Sub2 complex dimer | | Descriptor: | BJ4_G0025130.mRNA.1.CDS.1, EM14S01-3B_G0007820.mRNA.1.CDS.1, TEX1 isoform 1, ... | | Authors: | Schuller, S.K, Schuller, J.M, Prabu, R.J, Baumgartner, M, Bonneau, F, basquin, J, Conti, E. | | Deposit date: | 2020-10-22 | | Release date: | 2020-12-02 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Structural insights into the nucleic acid remodeling mechanisms of the yeast THO-Sub2 complex.
Elife, 9, 2020
|
|
7APX
 
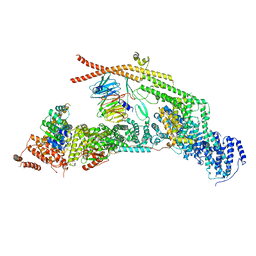 | | yeast THO-Sub2 complex | | Descriptor: | ATP-dependent RNA helicase SUB2, Protein TEX1, THO complex subunit 2,Tho2, ... | | Authors: | Schuller, S.K, Schuller, J.M, Prabu, R.J, Baumgartner, M, Bonneau, F, basquin, J, Conti, E. | | Deposit date: | 2020-10-20 | | Release date: | 2020-12-02 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural insights into the nucleic acid remodeling mechanisms of the yeast THO-Sub2 complex.
Elife, 9, 2020
|
|
8P5O
 
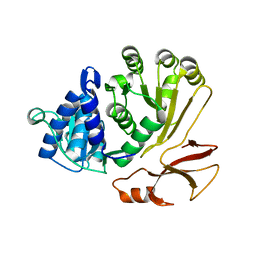 | | Proline activating adenylation domain of gramicidin S synthetase 2 - GrsB1-Acore | | Descriptor: | Gramicidin S synthase 2 | | Authors: | Stephan, P, Basquin, J, Caputi, L, O'Connor, S.E, Kries, H. | | Deposit date: | 2023-05-24 | | Release date: | 2023-07-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Directed Evolution of Piperazic Acid Incorporation by a Nonribosomal Peptide Synthetase.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
8PKI
 
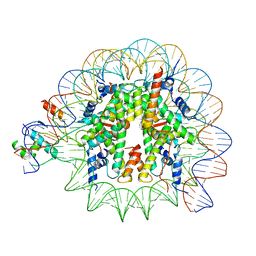 | | Cryo-EM structure of NR5A2-nucleosome complex SHL+5.5 | | Descriptor: | DNA, Histone H2A, Histone H2B type 1-C/E/G, ... | | Authors: | Kobayashi, W, Sappler, A, Bollschweiler, D, Kummecke, M, Basquin, J, Arslantas, E, Ruangroengkulrith, S, Hornberger, R, Duderstadt, K, Tachibana, K. | | Deposit date: | 2023-06-26 | | Release date: | 2024-02-28 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.58 Å) | | Cite: | Nucleosome-bound NR5A2 structure reveals pioneer factor mechanism by DNA minor groove anchor competition.
Nat.Struct.Mol.Biol., 2024
|
|
8PKJ
 
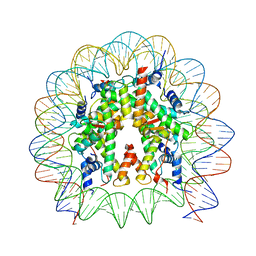 | | Cryo-EM structure of the nucleosome containing Nr5a2 motif at SHL+5.5 | | Descriptor: | DNA, Histone H2A, Histone H2B, ... | | Authors: | Kobayashi, W, Sappler, A, Bollschweiler, D, Kummecke, M, Basquin, J, Arslantas, E, Ruangroengkulrith, S, Hornberger, R, Duderstadt, K, Tachibana, K. | | Deposit date: | 2023-06-26 | | Release date: | 2024-02-28 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Nucleosome-bound NR5A2 structure reveals pioneer factor mechanism by DNA minor groove anchor competition.
Nat.Struct.Mol.Biol., 2024
|
|
5NKK
 
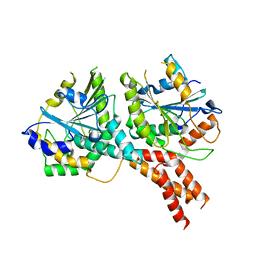 | | SMG8-SMG9 complex GDP bound | | Descriptor: | 1,2-ETHANEDIOL, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Li, L, Basquin, J, Conti, E. | | Deposit date: | 2017-03-31 | | Release date: | 2017-04-19 | | Last modified: | 2019-10-16 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Structure of a SMG8-SMG9 complex identifies a G-domain heterodimer in the NMD effector proteins.
RNA, 23, 2017
|
|
7NY7
 
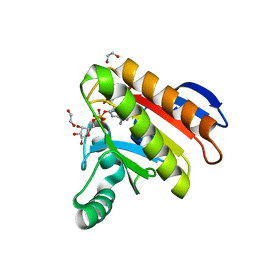 | | Crystal structure of the Capsaspora owczarzaki macroH2A macrodomain in complex with ADP-ribose | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5-DIPHOSPHORIBOSE, Histone macroH2A1.1 | | Authors: | Guberovic, I, Knobloch, G, Basquin, J, Buschbeck, M, Ladurner, A.G. | | Deposit date: | 2021-03-21 | | Release date: | 2021-11-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Evolution of a histone variant involved in compartmental regulation of NAD metabolism.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7NY6
 
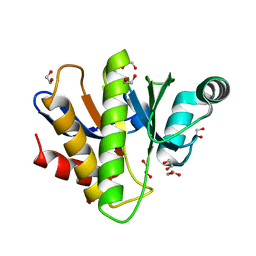 | | Crystal structure of the Capsaspora owczarzaki macroH2A macrodomain | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Histone macroH2A1.1 | | Authors: | Knobloch, G, Guberovic, I, Basquin, J, Buschbeck, M, Ladurner, A.G. | | Deposit date: | 2021-03-21 | | Release date: | 2021-11-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Evolution of a histone variant involved in compartmental regulation of NAD metabolism.
Nat.Struct.Mol.Biol., 28, 2021
|
|
6RO1
 
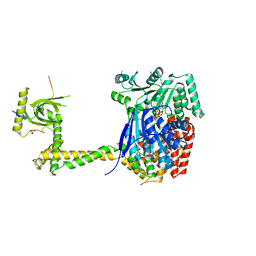 | | X-ray crystal structure of the MTR4 NVL complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, Exosome RNA helicase MTR4, ... | | Authors: | Lingaraju, M, Langer, L.M, Basquin, J, Falk, S, Conti, E. | | Deposit date: | 2019-05-10 | | Release date: | 2019-07-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.07 Å) | | Cite: | The MTR4 helicase recruits nuclear adaptors of the human RNA exosome using distinct arch-interacting motifs.
Nat Commun, 10, 2019
|
|
6RSX
 
 | |
6YUN
 
 | |
6ZCO
 
 | | Crystal Structure of C-terminal Dimerization Domain of Nucleocapsid Phosphoprotein from SARS-CoV-2, crystal form II | | Descriptor: | Nucleoprotein | | Authors: | Zinzula, L, Basquin, J, Nagy, I, Bracher, A. | | Deposit date: | 2020-06-11 | | Release date: | 2020-07-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.361 Å) | | Cite: | High-resolution structure and biophysical characterization of the nucleocapsid phosphoprotein dimerization domain from the Covid-19 severe acute respiratory syndrome coronavirus 2.
Biochem.Biophys.Res.Commun., 538, 2021
|
|
6SDK
 
 | |
6I59
 
 | | Long wavelength native-SAD phasing of Sen1 helicase | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-DIPHOSPHATE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Basu, S, Olieric, V, Matsugaki, N, Kawano, Y, Takashi, T, Huang, C.Y, Leonarski, F, Yamada, Y, Vera, L, Olieric, N, Basquin, J, Wojdyla, J.A, Diederichs, K, Yamamoto, M, Bunk, O, Wang, M. | | Deposit date: | 2018-11-13 | | Release date: | 2019-03-13 | | Last modified: | 2019-06-12 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Long-wavelength native-SAD phasing: opportunities and challenges.
Iucrj, 6, 2019
|
|
7QUJ
 
 | | Structure of NsNEPS2, a 7S-cis-trans nepetalactone synthase | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, NsNEPS2 | | Authors: | Hernandez Lozada, N.J, Hong, B, Wood, J.C, Caputi, L, Basquin, J, Chuang, L, Kunert, M, Rodriguez Lopez, C.R, Langley, C, Zhao, D, Buell, C.R, Lichman, B.R, O'Connor, S.E. | | Deposit date: | 2022-01-18 | | Release date: | 2022-12-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Biocatalytic routes to stereo-divergent iridoids.
Nat Commun, 13, 2022
|
|
5AIE
 
 | | Not4 ring domain in complex with Ubc4 | | Descriptor: | GENERAL NEGATIVE REGULATOR OF TRANSCRIPTION SUBUNIT 4, UBIQUITIN-CONJUGATING ENZYME E2 4, ZINC ION | | Authors: | Bhaskar, V, Basquin, J, Conti, E. | | Deposit date: | 2015-02-12 | | Release date: | 2015-04-29 | | Last modified: | 2015-05-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Architecture of the Ubiquitylation Module of the Yeast Ccr4-not Complex.
Structure, 23, 2015
|
|
5AJD
 
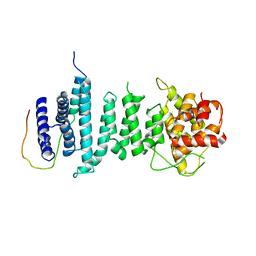 | |
5NNZ
 
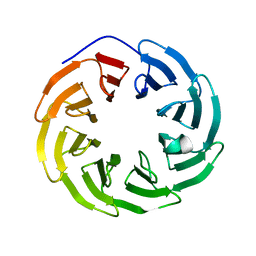 | | Crystal structure of human ODA16 | | Descriptor: | Dynein assembly factor with WDR repeat domains 1 | | Authors: | Lorentzen, E, Taschner, T, Basquin, J. | | Deposit date: | 2017-04-10 | | Release date: | 2017-06-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.651 Å) | | Cite: | Purification and crystal structure of human ODA16: Implications for ciliary import of outer dynein arms by the intraflagellar transport machinery.
Protein Sci., 29, 2020
|
|
1HK9
 
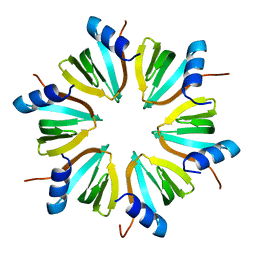 | |
1I5L
 
 | | CRYSTAL STRUCTURE OF AN SM-LIKE PROTEIN (AF-SM1) FROM ARCHAEOGLOBUS FULGIDUS COMPLEXED WITH SHORT POLY-U RNA | | Descriptor: | 5'-R(*UP*UP*U)-3', PUTATIVE SNRNP SM-LIKE PROTEIN AF-SM1, URIDINE | | Authors: | Toro, I, Thore, S, Mayer, C, Basquin, J, Seraphin, B, Suck, D. | | Deposit date: | 2001-02-28 | | Release date: | 2001-08-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | RNA binding in an Sm core domain: X-ray structure and functional analysis of an archaeal Sm protein complex.
EMBO J., 20, 2001
|
|
1I4K
 
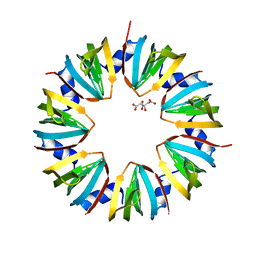 | | CRYSTAL STRUCTURE OF AN SM-LIKE PROTEIN (AF-SM1) FROM ARCHAEOGLOBUS FULGIDUS AT 2.5A RESOLUTION | | Descriptor: | CITRIC ACID, PUTATIVE SNRNP SM-LIKE PROTEIN | | Authors: | Toro, I, Thore, S, Mayer, C, Basquin, J, Seraphin, B, Suck, D. | | Deposit date: | 2001-02-22 | | Release date: | 2001-08-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | RNA binding in an Sm core domain: X-ray structure and functional analysis of an archaeal Sm protein complex.
EMBO J., 20, 2001
|
|
8DQ6
 
 | |
5MZN
 
 | | Helicase Sen1 | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-DIPHOSPHATE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Leonaite, B, Basquin, J, Conti, E. | | Deposit date: | 2017-02-01 | | Release date: | 2017-04-19 | | Last modified: | 2017-06-14 | | Method: | X-RAY DIFFRACTION (1.787 Å) | | Cite: | Sen1 has unique structural features grafted on the architecture of the Upf1-like helicase family.
EMBO J., 36, 2017
|
|
5MZH
 
 | | Crystal structure of ODA16 from Chlamydomonas reinhardtii | | Descriptor: | Dynein assembly factor with WDR repeat domains 1, SULFATE ION | | Authors: | Lorentzen, E, Taschner, M, Basquin, J, Mourao, A. | | Deposit date: | 2017-01-31 | | Release date: | 2017-03-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.398 Å) | | Cite: | Structural basis of outer dynein arm intraflagellar transport by the transport adaptor protein ODA16 and the intraflagellar transport protein IFT46.
J. Biol. Chem., 292, 2017
|
|
