2ALV
 
 | | X-ray structural analysis of SARS coronavirus 3CL proteinase in complex with designed anti-viral inhibitors | | Descriptor: | N-((3S,6R)-6-((S,E)-4-ETHOXYCARBONYL-1-((S)-2-OXOPYRROLIDIN-3-YL)BUT-3-EN-2-YLCARBAMOYL)-2,9-DIMETHYL-4-OXODEC-8-EN-3-YL)-5-METHYLISOXAZOLE-3-CARBOXAMIDE, Replicase polyprotein 1ab | | Authors: | Ghosh, A.K, Xi, K, Ratia, K, Santarsiero, B.D, Fu, W, Harcourt, B.H, Rota, P.A, Baker, S.C, Johnson, M.E, Mesecar, A.D. | | Deposit date: | 2005-08-08 | | Release date: | 2006-08-08 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Design and synthesis of peptidomimetic severe acute respiratory syndrome chymotrypsin-like protease inhibitors.
J.Med.Chem., 48, 2005
|
|
3V3M
 
 | | Severe Acute Respiratory Syndrome Coronavirus (SARS-CoV) 3CL Protease in Complex with N-[(1R)-2-(tert-butylamino)-2-oxo-1-(pyridin-3-yl)ethyl]-N-(4-tert-butylphenyl)furan-2-carboxamide inhibitor. | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE, N-[(1R)-2-(tert-butylamino)-2-oxo-1-(pyridin-3-yl)ethyl]-N-(4-tert-butylphenyl)furan-2-carboxamide | | Authors: | Jacobs, J, Grum-Tokars, V, Zhou, Y, Turlington, M, Saldanha, S.A, Chase, P, Eggler, A, Dawson, E.S, Baez-Santos, Y.M, Tomar, S, Mielech, A.M, Baker, S.C, Lindsley, C.W, Hodder, P, Mesecar, A, Stauffer, S.R. | | Deposit date: | 2011-12-13 | | Release date: | 2013-01-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Discovery, Synthesis, And Structure-Based Optimization of a Series of N-(tert-Butyl)-2-(N-arylamido)-2-(pyridin-3-yl) Acetamides (ML188) as Potent Noncovalent Small Molecule Inhibitors of the Severe Acute Respiratory Syndrome Coronavirus (SARS-CoV) 3CL Protease.
J.Med.Chem., 56, 2013
|
|
4YPT
 
 | | X-ray structural of three tandemly linked domains of nsp3 from murine hepatitis virus at 2.60 Angstroms resolution | | Descriptor: | GLYCEROL, Replicase polyprotein 1ab, ZINC ION | | Authors: | Chen, Y, Savinov, S.N, Mielech, A.M, Cao, T, Baker, S.C, Mesecar, A.D. | | Deposit date: | 2015-03-13 | | Release date: | 2015-08-26 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.6009 Å) | | Cite: | X-ray Structural and Functional Studies of the Three Tandemly Linked Domains of Non-structural Protein 3 (nsp3) from Murine Hepatitis Virus Reveal Conserved Functions.
J.Biol.Chem., 290, 2015
|
|
4MM3
 
 | |
2FE8
 
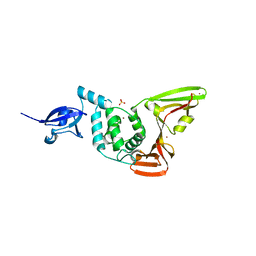 | | SARS coronavirus papain-like protease: structure of a viral deubiquitinating enzyme | | Descriptor: | BROMIDE ION, Replicase polyprotein 1ab, SULFATE ION, ... | | Authors: | Ratia, K, Santarsiero, B.D, Mesecar, A.D. | | Deposit date: | 2005-12-15 | | Release date: | 2006-03-21 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Severe acute respiratory syndrome coronavirus papain-like protease: Structure of a viral deubiquitinating enzyme
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
3MJ5
 
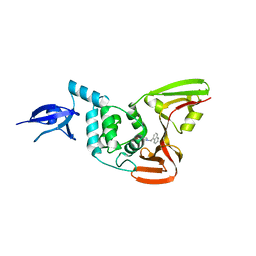 | | Severe Acute Respiratory Syndrome-Coronavirus Papain-Like Protease Inhibitors: Design, Synthesis, Protein-Ligand X-ray Structure and Biological Evaluation | | Descriptor: | N-(1,3-benzodioxol-5-ylmethyl)-1-[(1R)-1-naphthalen-1-ylethyl]piperidine-4-carboxamide, Replicase polyprotein 1a, ZINC ION | | Authors: | Mesecar, A.D, Ratia, K.M, Pegan, S.D. | | Deposit date: | 2010-04-12 | | Release date: | 2010-06-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.63 Å) | | Cite: | Severe acute respiratory syndrome coronavirus papain-like novel protease inhibitors: design, synthesis, protein-ligand X-ray structure and biological evaluation
J.Med.Chem., 53, 2010
|
|
5KO3
 
 | |
4OW0
 
 | |
4OVZ
 
 | |
7JU7
 
 | | The crystal structure of SARS-CoV-2 Main Protease in complex with masitinib | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Tan, K, Maltseva, N.I, Welk, L.F, Jedrzejczak, R.P, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-08-19 | | Release date: | 2020-09-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Masitinib is a broad coronavirus 3CL inhibitor that blocks replication of SARS-CoV-2.
Science, 373, 2021
|
|
7L5D
 
 | | The crystal structure of SARS-CoV-2 Main Protease in complex with demethylated analog of masitinib | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Tan, K, Maltseva, N.I, Jedrzejczak, R.P, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-12-21 | | Release date: | 2020-12-30 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Masitinib is a broad coronavirus 3CL inhibitor that blocks replication of SARS-CoV-2.
Science, 373, 2021
|
|
2QBZ
 
 | |
2QIQ
 
 | |
1E2R
 
 | | CYTOCHROME CD1 NITRITE REDUCTASE, REDUCED AND CYANIDE BOUND | | Descriptor: | CYANIDE ION, GLYCEROL, HEME C, ... | | Authors: | Fulop, V. | | Deposit date: | 2000-05-24 | | Release date: | 2000-05-30 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | X-Ray Crystallographic Study of Cyanide Binding Provides Insights Into the Structure-Function Relationship for Cytochrome Cd1 Nitrite Reductase from Paracoccus Pantotrophus.
J.Biol.Chem., 275, 2000
|
|
1QKS
 
 | | CYTOCHROME CD1 NITRITE REDUCTASE, OXIDISED FORM | | Descriptor: | CYTOCHROME CD1 NITRITE REDUCTASE, GLYCEROL, HEME C, ... | | Authors: | Fulop, V. | | Deposit date: | 1999-08-05 | | Release date: | 1999-08-18 | | Last modified: | 2019-10-09 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | The anatomy of a bifunctional enzyme: structural basis for reduction of oxygen to water and synthesis of nitric oxide by cytochrome cd1.
Cell(Cambridge,Mass.), 81, 1995
|
|
